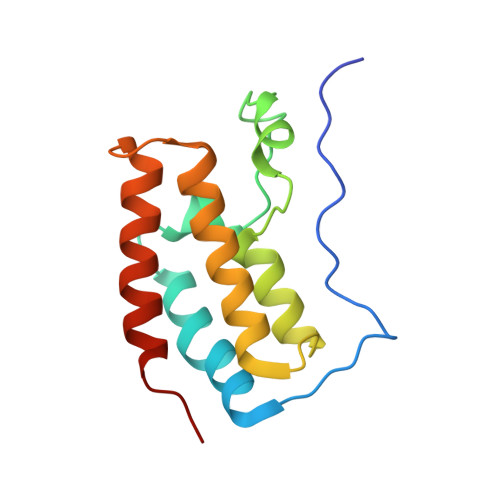
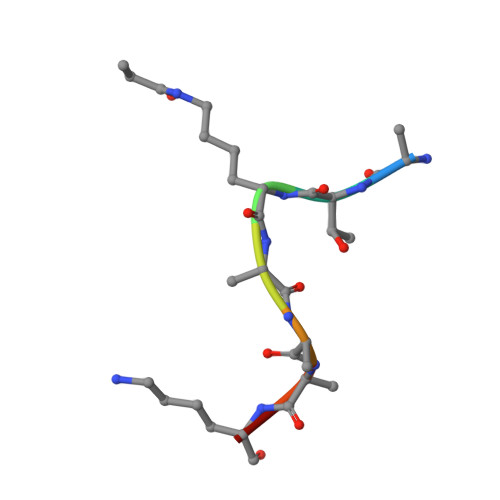
Interaction of propionylated and butyrylated histone H3 lysine marks with Brd4 bromodomains
Vollmuth, F., Geyer, M.(2010) Angew Chem Int Ed Engl 49: 6768-6772
- PubMed: 20715035
- DOI: https://doi.org/10.1002/anie.201002724
- Primary Citation of Related Structures:
3MUK, 3MUL - Abteilung Physikalische Biochemie, Max-Planck-Institut für molekulare Physiologie, Otto-Hahn-Strasse 11, 44227 Dortmund, Germany.
Organizational Affiliation: