Structural Characterization of HBXIP: The Protein That Interacts with the Anti-Apoptotic Protein Survivin and the Oncogenic Viral Protein HBx.
Garcia-Saez, I., Lacroix, F.B., Blot, D., Gabel, F., Skoufias, D.A.(2011) J Mol Biology 405: 331-340
- PubMed: 21059355
- DOI: https://doi.org/10.1016/j.jmb.2010.10.046
- Primary Citation of Related Structures:
3MS6, 3MSH - PubMed Abstract:
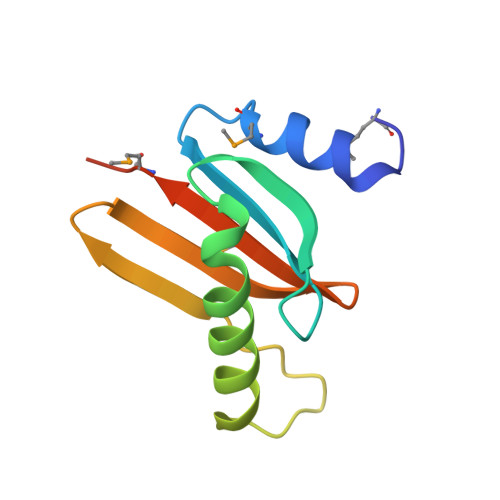
Hepatitis B X-interacting protein (HBXIP) is a ubiquitous protein that was originally identified as a binding partner of the hepatitis B viral protein HBx. HBXIP is also thought to serve as an anti-apoptotic cofactor of survivin, promoting the suppression of pro-caspase-9 activation. Here were port the crystal structure of the shortest isoform of HBXIP (91 aa long,∼11 kDa) at 1.5 Å resolution. HBXIP crystal shows a monomer per asymmetric unit, with a profilin-like fold which is common to a super family of proteins, the Roadblock/LC7 domain family involved in protein-protein interactions. Based on this fold, we propose that HBXIP can form a dimer that can indeed be found in the crystal when symmetric molecules are generated around the asymmetric unit. This dimer shows an extended β-sheet area formed by 10 anti-parallel β-strands from both subunits. Another interesting aspect of the proposed HBXIP dimer interface is the presence of a small leucine zipper between the two α2 helices of each monomer. In solution, the scattering curve obtained by small-angle X-ray scattering for the sample used for crystallization indicates that the protein is dimeric form in solution. The fit between the experimental small angle X-ray scattering curve and the back calculated curves for two potential crystal dimers shows a significant preference for the Roadblock/LC7 fold dimer model. Moreover, the HBXIP crystal structure represents a step towards understanding the cellular role of HBXIP.
- Institut de Biologie Structurale J.-P. Ebel, UMR 5075, Grenoble Cedex 1, France. isabel.garcia@ibs.fr
Organizational Affiliation: