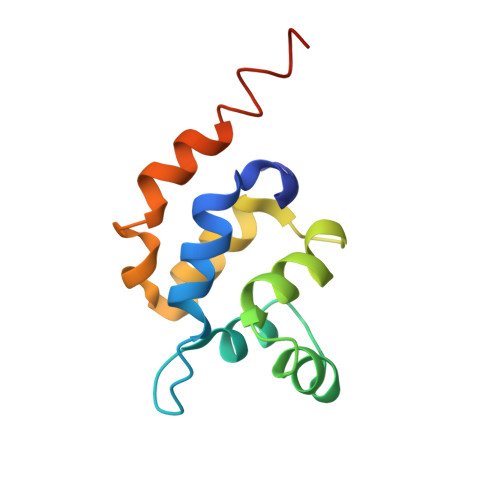
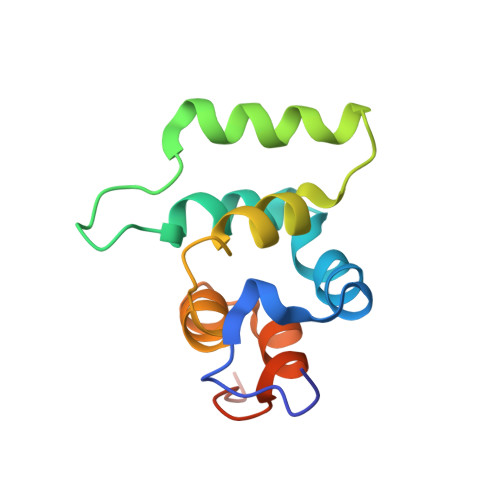
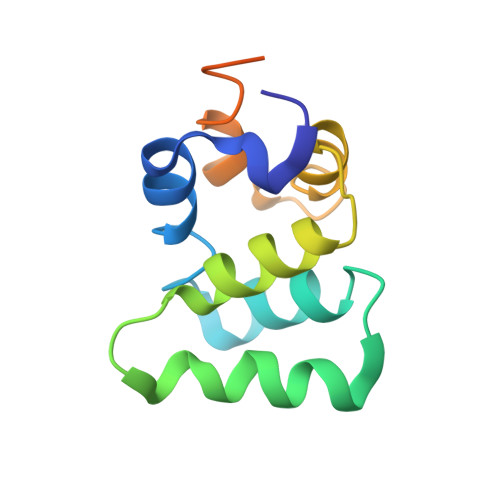
Helical assembly in the MyD88-IRAK4-IRAK2 complex in TLR/IL-1R signalling.
Lin, S.C., Lo, Y.C., Wu, H.(2010) Nature 465: 885-890
- PubMed: 20485341
- DOI: https://doi.org/10.1038/nature09121
- Primary Citation of Related Structures:
3MOP - PubMed Abstract:
MyD88, IRAK4 and IRAK2 are critical signalling mediators of the TLR/IL1-R superfamily. Here we report the crystal structure of the MyD88-IRAK4-IRAK2 death domain (DD) complex, which surprisingly reveals a left-handed helical oligomer that consists of 6 MyD88, 4 IRAK4 and 4 IRAK2 DDs. Assembly of this helical signalling tower is hierarchical, in which MyD88 recruits IRAK4 and the MyD88-IRAK4 complex recruits the IRAK4 substrates IRAK2 or the related IRAK1. Formation of these Myddosome complexes brings the kinase domains of IRAKs into proximity for phosphorylation and activation. Composite binding sites are required for recruitment of the individual DDs in the complex, which are confirmed by mutagenesis and previously identified signalling mutations. Specificities in Myddosome formation are dictated by both molecular complementarity and correspondence of surface electrostatics. The MyD88-IRAK4-IRAK2 complex provides a template for Toll signalling in Drosophila and an elegant mechanism for versatile assembly and regulation of DD complexes in signal transduction.
- Department of Biochemistry, Weill Cornell Medical College, New York, New York 10021, USA.
Organizational Affiliation: