Crystallization and X-ray structure analysis of a thermostable penicillin G acylase from Alcaligenes faecalis.
Varshney, N.K., Kumar, R.S., Ignatova, Z., Prabhune, A., Pundle, A., Dodson, E., Suresh, C.G.(2012) Acta Crystallogr Sect F Struct Biol Cryst Commun 68: 273-277
- PubMed: 22442220
- DOI: https://doi.org/10.1107/S1744309111053930
- Primary Citation of Related Structures:
3K3W, 3ML0 - PubMed Abstract:
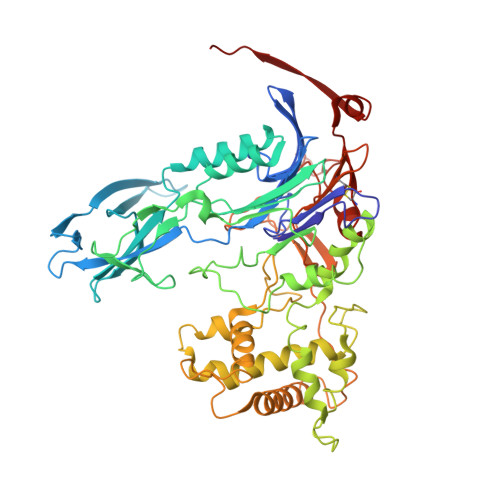
The enzyme penicillin G acylase (EC 3.5.1.11) catalyzes amide-bond cleavage in benzylpenicillin (penicillin G) to yield 6-aminopenicillanic acid, an intermediate chemical used in the production of semisynthetic penicillins. A thermostable penicillin G acylase from Alcaligenes faecalis (AfPGA) has been crystallized using the hanging-drop vapour-diffusion method in two different space groups: C222(1), with unit-cell parameters a = 72.9, b = 86.0, c = 260.2 , and P4(1)2(1)2, with unit-cell parameters a = b = 85.6, c = 298.8 . Data were collected at 293 and the structure was determined using the molecular-replacement method. Like other penicillin acylases, AfPGA belongs to the N-terminal nucleophilic hydrolase superfamily, has undergone post-translational processing and has a serine as the N-terminal residue of the β-chain. A disulfide bridge has been identified in the structure that was not found in the other two known penicillin G cylase structures. The presence of the disulfide bridge is perceived to be one factor that confers higher stability to this enzyme.
- Division of Biochemical Sciences, National Chemical Laboratory, Pune 411 008, India.
Organizational Affiliation: