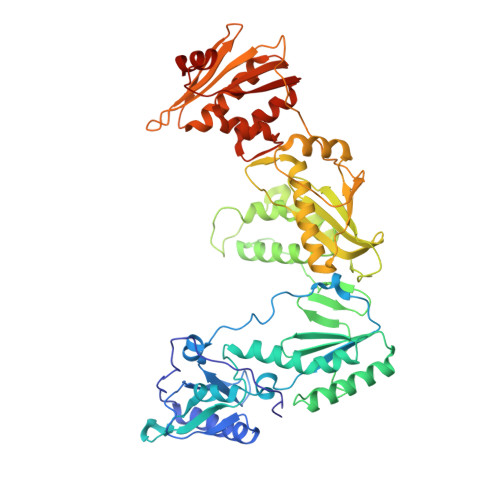
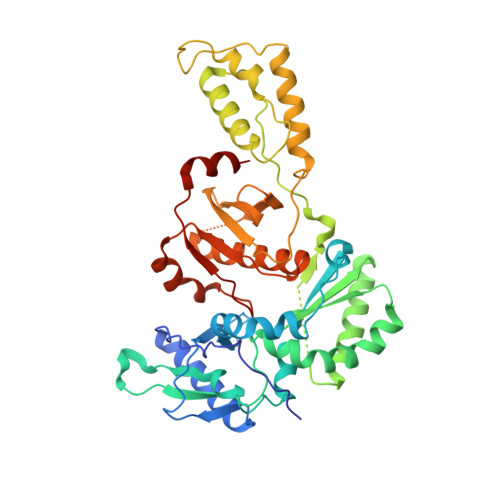
Crystal Structures of HIV-1 Reverse Transcriptase with Etravirine (TMC125) and Rilpivirine (TMC278): Implications for Drug Design.
Lansdon, E.B., Brendza, K.M., Hung, M., Wang, R., Mukund, S., Jin, D., Birkus, G., Kutty, N., Liu, X.(2010) J Med Chem 53: 4295-4299
- PubMed: 20438081
- DOI: https://doi.org/10.1021/jm1002233
- Primary Citation of Related Structures:
3MEC, 3MED, 3MEE, 3MEG - PubMed Abstract:
Diarylpyrimidine (DAPY) non-nucleoside reverse transcriptase inhibitors (NNRTIs) have inherent flexibility, helping to maintain activity against a wide range of resistance mutations. Crystal structures were determined with wild-type and K103N HIV-1 reverse transcriptase with etravirine (TMC125) and rilpivirine (TMC278). These structures reveal a similar binding mode for TMC125 and TMC278, whether bound to wild-type or K103N RT. Comparison to previously published structures reveals differences in binding modes for TMC125 and differences in protein conformation for TMC278.
- Gilead Sciences, Inc., 333 Lakeside Drive, Foster City, California 94404, USA. eric.lansdon@gilead.com
Organizational Affiliation: