Crystal structure of the ternary complex of full length centaurin alpha-1, KIF13B FHA domain, and IP4
Shen, L., Tong, Y., Tempel, W., MacKenzie, F., Arrowsmith, C.H., Edwards, A.M., Bountra, C., Weigelt, J., Bochkarev, A., Park, H.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Kinesin-like protein KIF13B | 124 | Homo sapiens | Mutation(s): 0 Gene Names: KIF13B, GAKIN, KIAA0639 | 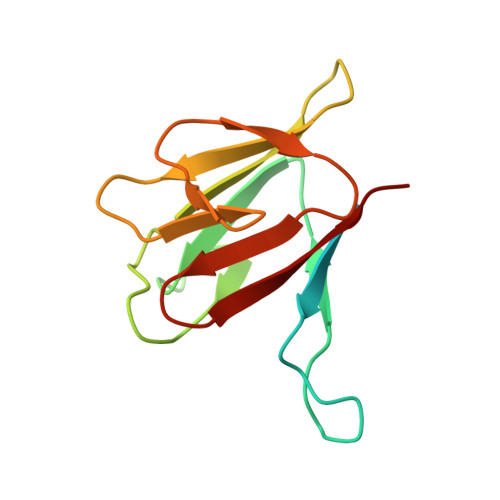 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q9NQT8 (Homo sapiens) Explore Q9NQT8 Go to UniProtKB: Q9NQT8 | |||||
PHAROS: Q9NQT8 GTEx: ENSG00000197892 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9NQT8 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Arf-GAP with dual PH domain-containing protein 1 | 392 | Homo sapiens | Mutation(s): 0 Gene Names: ADAP1, CENTA1 | 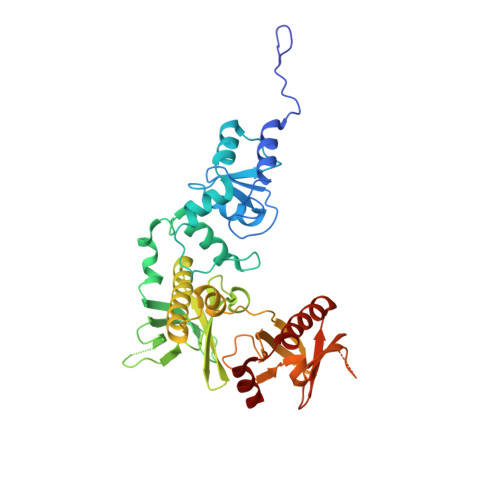 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for O75689 (Homo sapiens) Explore O75689 Go to UniProtKB: O75689 | |||||
PHAROS: O75689 GTEx: ENSG00000105963 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | O75689 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| IP9 Query on IP9 | I [auth C] | (2R)-3-{[(R)-{[(1S,2S,3R,4S,5S,6S)-2,6-dihydroxy-3,4,5-tris(phosphonooxy)cyclohexyl]oxy}(hydroxy)phosphoryl]oxy}propane
-1,2-diyl dioctanoate C25 H50 O22 P4 ANFYVAHJWGJYAT-QLCNXWICSA-N |  | ||
| ZN Query on ZN | H [auth C], M [auth D] | ZINC ION Zn PTFCDOFLOPIGGS-UHFFFAOYSA-N |  | ||
| UNX Query on UNX | E [auth A] F [auth A] G [auth B] J [auth C] K [auth C] | UNKNOWN ATOM OR ION X |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 115.797 | α = 90 |
| b = 115.797 | β = 90 |
| c = 189.269 | γ = 90 |
| Software Name | Purpose |
|---|---|
| DENZO | data reduction |
| SCALEPACK | data scaling |
| REFMAC | refinement |
| PDB_EXTRACT | data extraction |
| HKL-2000 | data scaling |