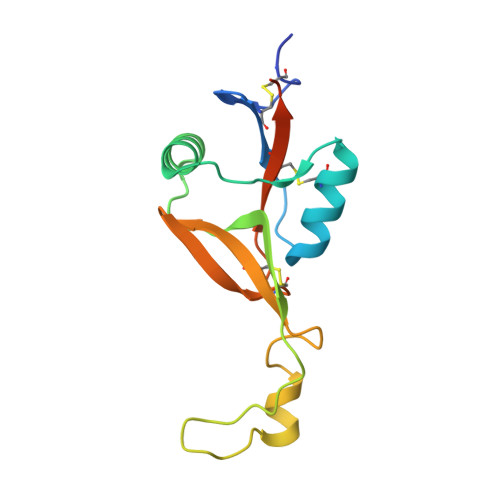
Molecular architecture of mouse activating NKR-P1 receptors.
Kolenko, P., Rozbesky, D., Vanek, O., Kopecky, V., Hofbauerova, K., Novak, P., Pompach, P., Hasek, J., Skalova, T., Bezouska, K., Dohnalek, J.(2011) J Struct Biol 175: 434-441
- PubMed: 21600988
- DOI: https://doi.org/10.1016/j.jsb.2011.05.001
- Primary Citation of Related Structures:
3M9Z - PubMed Abstract:
Receptors belonging to NKR-P1 family and their specific Clr ligands form an alternative missing self recognition system critical in immunity against tumors and viruses, elimination of tumor cells subjected to genotoxic stress, activation of T cell dependent immune response, and hypertension. The three-dimensional structure of the extracellular domain of the mouse natural killer (NK) cell receptor mNKR-P1Aex has been determined by X-ray diffraction. The core of the C-type lectin domain (CTLD) is homologous to the other CTLD receptors whereas one quarter of the domain forms an extended loop interacting tightly with a neighboring loop in the crystal. This domain swapping mechanism results in a compact interaction interface. A second dimerization interface resembles the known arrangement of other CTLD NK receptors. A functional dimeric form of the receptor is suggested, with the loop, evolutionarily conserved within this family, proposed to participate in interactions with ligands.
- Institute of Macromolecular Chemistry, v.v.i., Academy of Sciences of the Czech Republic, Heyrovského náměstí 2, 16206 Prague 6, Czech Republic.
Organizational Affiliation: