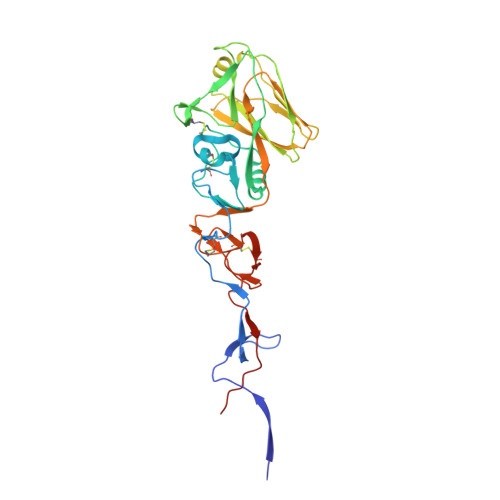
Structures of receptor complexes of a North American H7N2 influenza hemagglutinin with a loop deletion in the receptor binding site.
Yang, H., Chen, L.M., Carney, P.J., Donis, R.O., Stevens, J.(2010) PLoS Pathog 6: e1001081-e1001081
- PubMed: 20824086
- DOI: https://doi.org/10.1371/journal.ppat.1001081
- Primary Citation of Related Structures:
3M5G, 3M5H, 3M5I, 3M5J - PubMed Abstract:
Human infections with subtype H7 avian influenza viruses have been reported as early as 1979. In 1996, a genetically stable 24-nucleotide deletion emerged in North American H7 influenza virus hemagglutinins, resulting in an eight amino acid deletion in the receptor-binding site. The continuous circulation of these viruses in live bird markets, as well as its documented ability to infect humans, raises the question of how these viruses achieve structural stability and functionality. Here we report a detailed molecular analysis of the receptor binding site of the North American lineage subtype H7N2 virus A/New York/107/2003 (NY107), including complexes with an avian receptor analog (3'-sialyl-N-acetyllactosamine, 3'SLN) and two human receptor analogs (6'-sialyl-N-acetyllactosamine, 6'SLN; sialyllacto-N-tetraose b, LSTb). Structural results suggest a novel mechanism by which residues Arg220 and Arg229 (H3 numbering) are used to compensate for the deletion of the 220-loop and form interactions with the receptor analogs. Glycan microarray results reveal that NY107 maintains an avian-type (alpha2-3) receptor binding profile, with only moderate binding to human-type (alpha2-6) receptor. Thus despite its dramatically altered receptor binding site, this HA maintains functionality and confirms a need for continued influenza virus surveillance of avian and other animal reservoirs to define their zoonotic potential.
- Influenza Division, Centers for Disease Control and Prevention, Atlanta, Georgia, USA.
Organizational Affiliation: