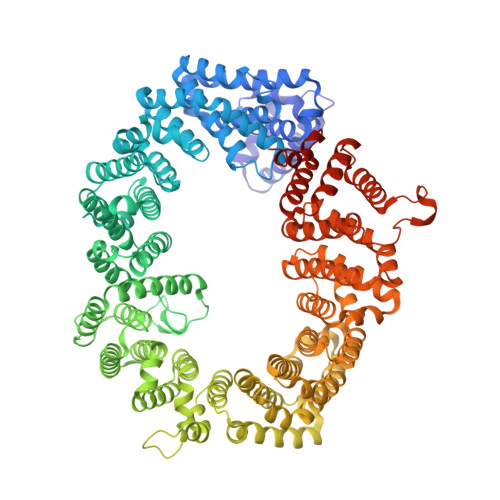
An allosteric mechanism to displace nuclear export cargo from CRM1 and RanGTP by RanBP1
Koyama, M., Matsuura, Y.(2010) EMBO J 29: 2002-2013
- PubMed: 20485264
- DOI: https://doi.org/10.1038/emboj.2010.89
- Primary Citation of Related Structures:
3M1I - PubMed Abstract:
The karyopherin CRM1 mediates nuclear export of proteins and ribonucleoproteins bearing a leucine-rich nuclear export signal (NES). To elucidate the precise mechanism by which NES-cargos are dissociated from CRM1 in the cytoplasm, which is important for transport directionality, we determined a 2.0-A resolution crystal structure of yeast CRM1:RanBP1:RanGTP complex, an intermediate in the disassembly of the CRM1 nuclear export complex. The structure shows that on association of Ran-binding domain (RanBD) of RanBP1 with CRM1:NES-cargo:RanGTP complex, RanBD and the C-terminal acidic tail of Ran induce a large movement of the intra-HEAT9 loop of CRM1. The loop moves to the CRM1 inner surface immediately behind the NES-binding site and causes conformational rearrangements in HEAT repeats 11 and 12 so that the hydrophobic NES-binding cleft on the CRM1 outer surface closes, squeezing out the NES-cargo. This allosteric mechanism accelerates dissociation of NES by over two orders of magnitude. Structure-based mutagenesis indicated that the HEAT9 loop also functions as an allosteric autoinhibitor to stabilize CRM1 in a conformation that is unable to bind NES-cargo in the absence of RanGTP.
- Division of Biological Science, Graduate School of Science, Nagoya University, Furo-cho, Chikusa-ku, Nagoya, Japan.
Organizational Affiliation: