Replication through an abasic DNA lesion: structural basis for adenine selectivity
Obeid, S., Blatter, N., Kranaster, R., Schnur, A., Diederichs, K., Welte, W., Marx, A.(2010) EMBO J 29: 1738-1747
- PubMed: 20400942
- DOI: https://doi.org/10.1038/emboj.2010.64
- Primary Citation of Related Structures:
3LWL, 3LWM - PubMed Abstract:
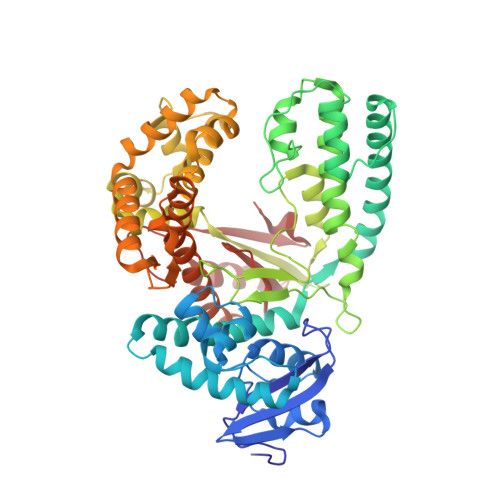
Abasic sites represent the most frequent DNA lesions in the genome that have high mutagenic potential and lead to mutations commonly found in human cancers. Although these lesions are devoid of the genetic information, adenine is most efficiently inserted when abasic sites are bypassed by DNA polymerases, a phenomenon termed A-rule. In this study, we present X-ray structures of a DNA polymerase caught while incorporating a nucleotide opposite an abasic site. We found that a functionally important tyrosine side chain directs for nucleotide incorporation rather than DNA. It fills the vacant space of the absent template nucleobase and thereby mimics a pyrimidine nucleobase directing for preferential purine incorporation opposite abasic residues because of enhanced geometric fit to the active site. This amino acid templating mechanism was corroborated by switching to pyrimidine specificity because of mutation of the templating tyrosine into tryptophan. The tyrosine is located in motif B and highly conserved throughout evolution from bacteria to humans indicating a general amino acid templating mechanism for bypass of non-instructive lesions by DNA polymerases at least from this sequence family.
- Konstanz Research School Chemical Biology, University of Konstanz, Konstanz, Germany.
Organizational Affiliation: