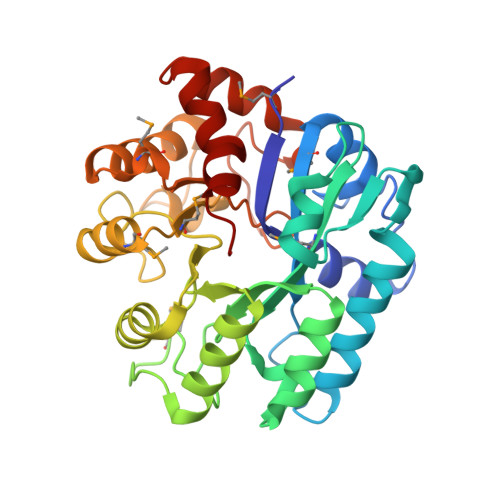
Structure of lmo2462, a Listeria monocytogenes amidohydrolase family putative dipeptidase
Anderson, S.M., Wawrzak, Z., Onopriyenko, O., Hasseman, J., Edwards, A., Savchenko, A., Anderson, W.F., Center for Structural Genomics of Infectious Diseases (CSGID)To be published.