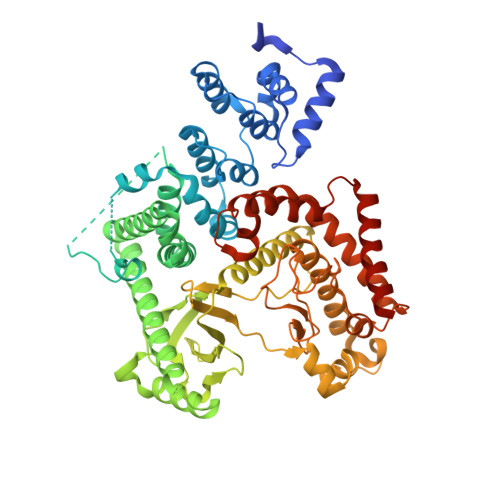
Crystal structure of human PIK3C3 in complex with 3-[4-(4-Morpholinyl)thieno[3,2-d]pyrimidin-2-yl]-phenol
Tresaugues, L., Welin, M., Arrowsmith, C.H., Berglund, H., Bountra, C., Collins, R., Edwards, A.M., Flodin, S., Flores, A., Graslund, S., Hammarstrom, M., Johansson, I., Karlberg, T., Kotenyova, T., Kraulis, P., Moche, M., Nyman, T., Persson, C., Schuler, H., Schutz, P., Siponen, M.I., Thorsell, A.G., Van den Berg, S., Wahlberg, E., Weigelt, J., Wisniewska, M., Nordlund, P.To be published.