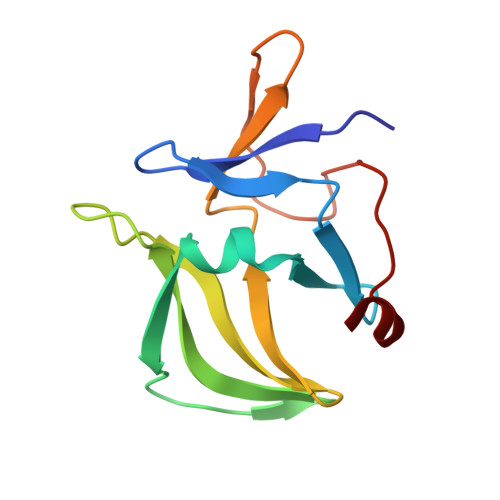
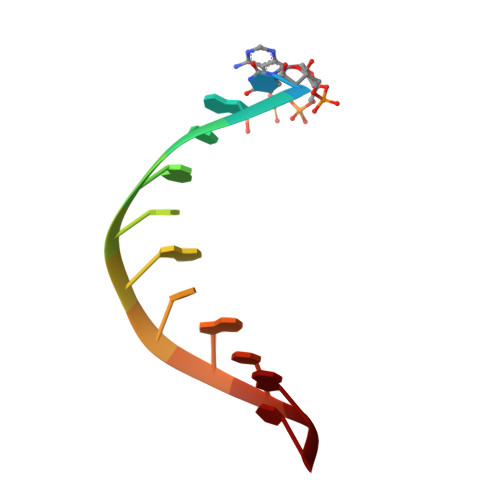
The Structural Basis of 5' Triphosphate Double-Stranded RNA Recognition by RIG-I C-Terminal Domain.
Lu, C., Xu, H., Ranjith-Kumar, C.T., Brooks, M.T., Hou, T.Y., Hu, F., Herr, A.B., Strong, R.K., Kao, C.C., Li, P.(2010) Structure 18: 1032-1043
- PubMed: 20637642
- DOI: https://doi.org/10.1016/j.str.2010.05.007
- Primary Citation of Related Structures:
3LRN, 3LRR - PubMed Abstract:
RIG-I is a cytosolic sensor of viral RNA that plays crucial roles in the induction of type I interferons. The C-terminal domain (CTD) of RIG-I is responsible for the recognition of viral RNA with 5' triphosphate (ppp). However, the mechanism of viral RNA recognition by RIG-I is still not fully understood. Here, we show that RIG-I CTD binds 5' ppp dsRNA or ssRNA, as well as blunt-ended dsRNA, and exhibits the highest affinity for 5' ppp dsRNA. Crystal structures of RIG-I CTD bound to 5' ppp dsRNA with GC- and AU-rich sequences revealed that RIG-I recognizes the termini of the dsRNA and interacts with the 5' ppp through extensive electrostatic interactions. Mutagenesis and RNA-binding studies demonstrated that similar binding surfaces are involved in the recognition of different forms of RNA. Mutations of key residues at the RNA-binding surface affected RIG-I signaling in cells.
- Department of Biochemistry and Biophysics, Texas A&M University, College Station, TX 77843-2128, USA.
Organizational Affiliation: