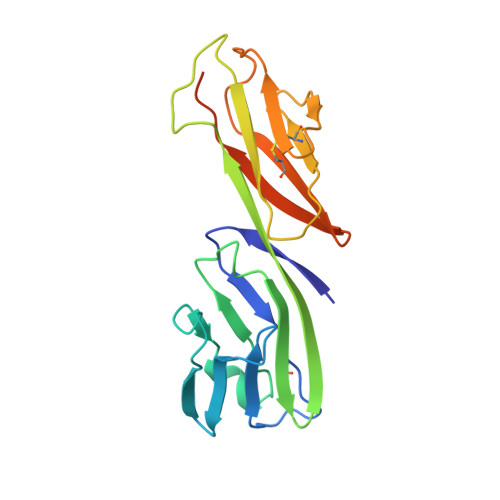
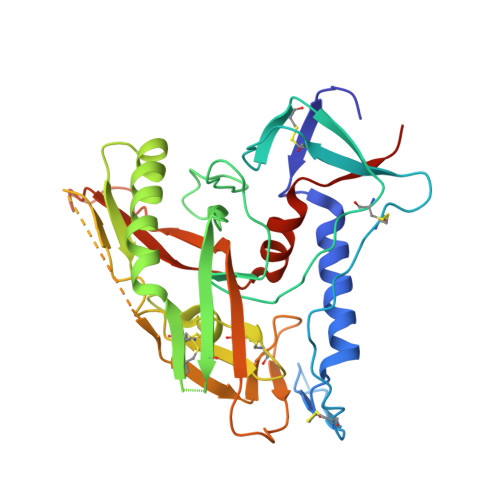
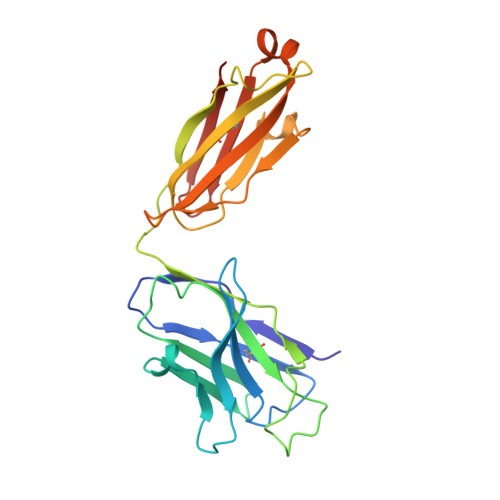
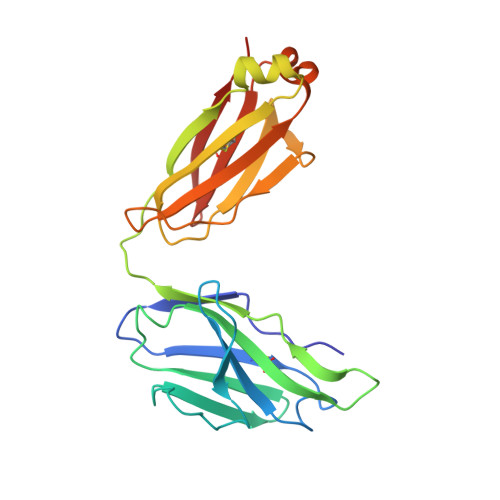
Structure of a clade C HIV-1 gp120 bound to CD4 and CD4-induced antibody reveals anti-CD4 polyreactivity.
Diskin, R., Marcovecchio, P.M., Bjorkman, P.J.(2010) Nat Struct Mol Biol 17: 608-613
- PubMed: 20357769
- DOI: https://doi.org/10.1038/nsmb.1796
- Primary Citation of Related Structures:
3LMJ, 3LQA - PubMed Abstract:
Strategies to combat HIV-1 require structural knowledge of envelope proteins from viruses in HIV-1 clade C, the most rapidly spreading subtype in the world. We present a crystal structure containing a clade C gp120 envelope. The structure, a complex between gp120, the host receptor CD4 and the CD4-induced antibody 21c, reveals that the 21c epitope involves contacts with gp120, a nonself antigen, and with CD4, an autoantigen. Binding studies using wild-type and mutant CD4 show that 21c Fab binds CD4 in the absence of gp120, and that binding of 21c to clade C and HIV-2 gp120s requires the crystallographically observed 21c-CD4 interaction. Additional binding data suggest a role for the gp120 V1V2 loop in creating a high-affinity, but slow-forming, epitope for 21c after CD4 binds. These results contribute to a molecular understanding of CD4-induced antibodies and provide the first visualization to our knowledge of a potentially autoreactive antibody Fab complexed with both self and nonself antigens.
- Division of Biology, California Institute of Technology, Pasadena, California, USA.
Organizational Affiliation: