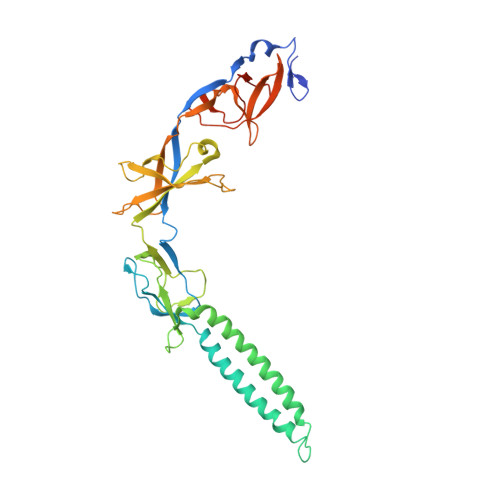
Metal-induced conformational changes in ZneB suggest an active role of membrane fusion proteins in efflux resistance systems.
De Angelis, F., Lee, J.K., O'Connell, J.D., Miercke, L.J., Verschueren, K.H., Srinivasan, V., Bauvois, C., Govaerts, C., Robbins, R.A., Ruysschaert, J.M., Stroud, R.M., Vandenbussche, G.(2010) Proc Natl Acad Sci U S A 107: 11038-11043
- PubMed: 20534468
- DOI: https://doi.org/10.1073/pnas.1003908107
- Primary Citation of Related Structures:
3LNN - PubMed Abstract:
Resistance nodulation cell division (RND)-based efflux complexes mediate multidrug and heavy-metal resistance in many Gram-negative bacteria. Efflux of toxic compounds is driven by membrane proton/substrate antiporters (RND protein) in the plasma membrane, linked by a membrane fusion protein (MFP) to an outer-membrane protein. The three-component complex forms an efflux system that spans the entire cell envelope. The MFP is required for the assembly of this complex and is proposed to play an important active role in substrate efflux. To better understand the role of MFPs in RND-driven efflux systems, we chose ZneB, the MFP component of the ZneCAB heavy-metal efflux system from Cupriavidus metallidurans CH34. ZneB is shown to be highly specific for Zn(2+) alone. The crystal structure of ZneB to 2.8 A resolution defines the basis for metal ion binding in the coordination site at a flexible interface between the beta-barrel and membrane proximal domains. The conformational differences observed between the crystal structures of metal-bound and apo forms are monitored in solution by spectroscopy and chromatography. The structural rearrangements between the two states suggest an active role in substrate efflux through metal binding and release.
- Laboratoire de Structure et Fonction des Membranes Biologiques, Faculté des Sciences, Centre de Biologie Structurale et de Bioinformatique, Université Libre de Bruxelles, B-1050 Brussels, Belgium.
Organizational Affiliation: