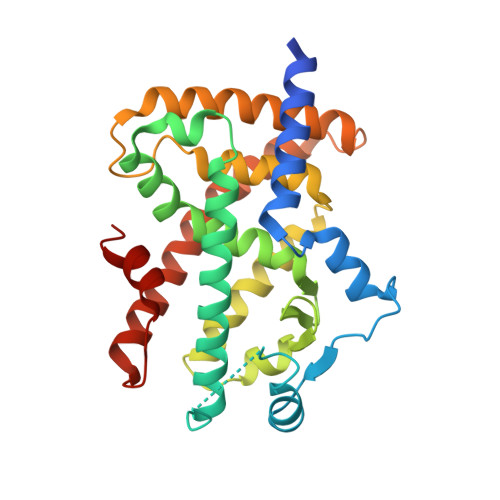
Discovery of a novel selective PPARgamma modulator from (-)-Cercosporamide derivatives
Furukawa, A., Arita, T., Satoh, S., Wakabayashi, K., Hayashi, S., Matsui, Y., Araki, K., Kuroha, M., Ohsumi, J.(2010) Bioorg Med Chem Lett 20: 2095-2098
- PubMed: 20219371
- DOI: https://doi.org/10.1016/j.bmcl.2010.02.073
- Primary Citation of Related Structures:
3LMP - PubMed Abstract:
In an investigation of (-)-Cercosporamide derivatives with a plasma glucose-lowering effect, we found that N-benzylcarboxamide derivative 4 was a partial agonist of PPARgamma. A SAR study of the substituents on carboxamide nitrogen afforded the N-(1-naphthyl)methylcarboxamide derivative 23 as the most potent selective PPARgamma modulator. An X-ray crystallography study revealed that compound 23 bounded to the PPARgamma ligand binding domain in a unique way without any interaction with helix12. Compound 23 displayed a potent plasma glucose-lowering effect in db/db mice without the undesirable increase in body fluid and heart weight that is typically observed when PPARgamma full agonists are administrated.
- Shinagawa R&D Center, Daiichi Sankyo Co., Ltd, 1-2-58, Hiromachi, Shinagawa-ku, Tokyo, Japan. furukawa.akihiro.zy@daiichisankyo.co.jp
Organizational Affiliation: