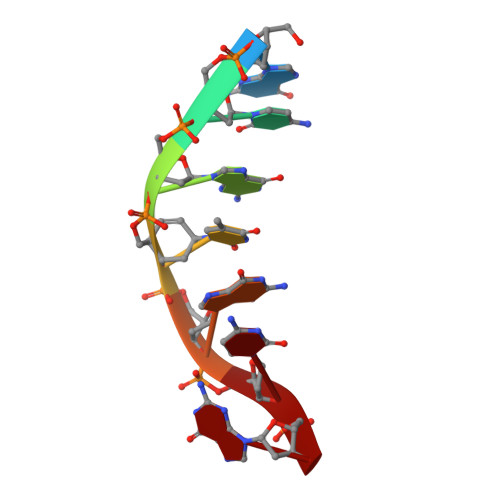
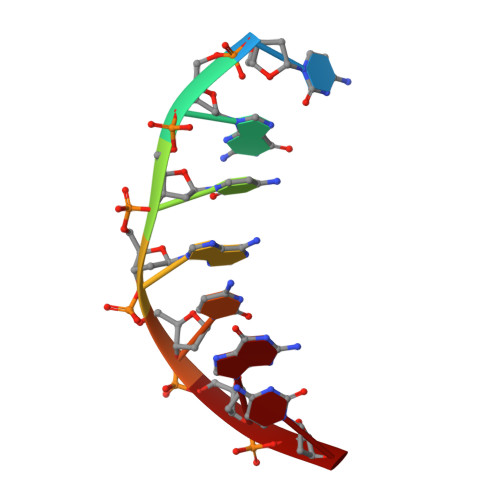
Comparison between the orthorhombic and tetragonal forms of the heptamer sequence d[GCG(xT)GCG]/d(CGCACGC)
Robeyns, K., Herdewijn, P., Van Meervelt, L.(2010) Acta Crystallogr Sect F Struct Biol Cryst Commun 66: 1028-1031
- PubMed: 20823518
- DOI: https://doi.org/10.1107/S1744309110031696
- Primary Citation of Related Structures:
3LLN - PubMed Abstract:
Cyclohexene nucleic acid (CeNA) building blocks can be introduced into natural DNA sequences without a large conformational influence because of the ability of the six-membered sugar ring to mimic both the C2'-endo and C3'-endo conformations of the naturally occurring ribofuranose sugar ring. The non-self-complementary DNA sequence d[GCG(xT)GCG]/d(CGCACGC) with one incorporated CeNA (xT) moiety crystallizes in two forms: orthorhombic and tetragonal. The tetragonal form, which diffracts to 3 A resolution, is a kinetically stable polymorph of the orthorhombic form [Robeyns et al. (2010), Artificial DNA, 1, 1-7], which diffracts to 1.17 A resolution and is the thermodynamically stable form of the CeNA-incorporated duplex. Here, the two structures are compared, with special emphasis on the differences in crystal packing and the irreversible conversion of the kinetic form into the high-resolution diffracting thermodynamic form.
- Department of Chemistry, Biomolecular Architecture and BioMacS, Katholieke Universiteit Leuven, Belgium.
Organizational Affiliation: