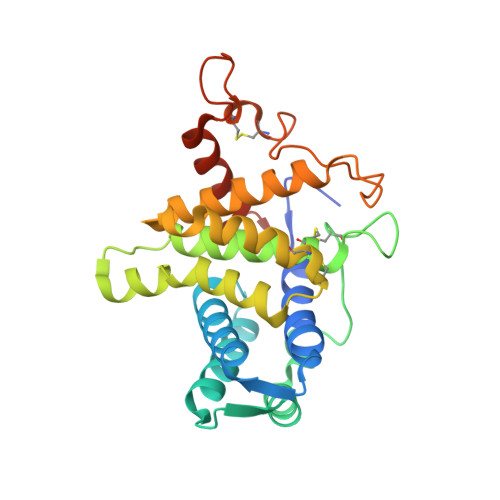
QSOX contains a pseudo-dimer of functional and degenerate sulfhydryl oxidase domains.
Alon, A., Heckler, E.J., Thorpe, C., Fass, D.(2010) FEBS Lett 584: 1521-1525
- PubMed: 20211621
- DOI: https://doi.org/10.1016/j.febslet.2010.03.001
- Primary Citation of Related Structures:
3LLI, 3LLK - PubMed Abstract:
Quiescin sulfhydryl oxidase (QSOX) catalyzes formation of disulfide bonds between cysteine residues in substrate proteins. Human QSOX1 is a multi-domain, monomeric enzyme containing a module related to the single-domain sulfhydryl oxidases of the Erv family. A partial QSOX1 crystal structure reveals a single-chain pseudo-dimer mimicking the quaternary structure of Erv enzymes. However, one pseudo-dimer "subunit" has lost its cofactor and catalytic activity. In QSOX evolution, a further concatenation to a member of the protein disulfide isomerase family resulted in an enzyme capable of both disulfide formation and efficient transfer to substrate proteins.
- Department of Structural Biology, Weizmann Institute of Science, Rehovot, Israel.
Organizational Affiliation: