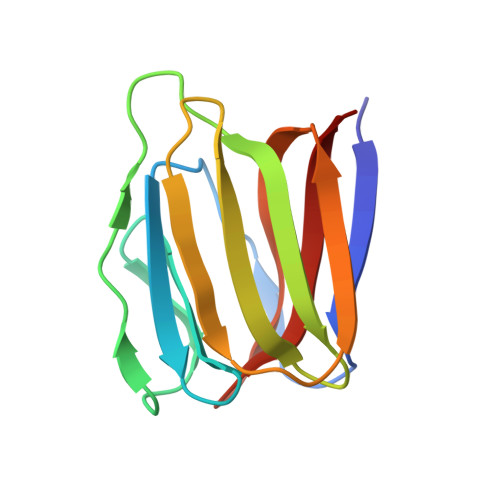
Monomerization of viral entry inhibitor griffithsin elucidates the relationship between multivalent binding to carbohydrates and anti-HIV activity.
Moulaei, T., Shenoy, S.R., Giomarelli, B., Thomas, C., McMahon, J.B., Dauter, Z., O'Keefe, B.R., Wlodawer, A.(2010) Structure 18: 1104-1115
- PubMed: 20826337
- DOI: https://doi.org/10.1016/j.str.2010.05.016
- Primary Citation of Related Structures:
3LKY, 3LL0, 3LL1, 3LL2 - PubMed Abstract:
Mutations were introduced to the domain-swapped homodimer of the antiviral lectin griffithsin (GRFT). Whereas several single and double mutants remained dimeric, insertion of either two or four amino acids at the dimerization interface resulted in a monomeric form of the protein (mGRFT). Monomeric character of the modified proteins was confirmed by sedimentation equilibrium ultracentrifugation and by their high resolution X-ray crystal structures, whereas their binding to carbohydrates was assessed by isothermal titration calorimetry. Cell-based antiviral activity assays utilizing different variants of mGRFT indicated that the monomeric form of the lectin had greatly reduced activity against HIV-1, suggesting that the antiviral activity of GRFT stems from crosslinking and aggregation of viral particles via multivalent interactions between GRFT and oligosaccharides present on HIV envelope glycoproteins. Atomic resolution crystal structure of a complex between mGRFT and nonamannoside revealed that a single mGRFT molecule binds to two different nonamannoside molecules through all three carbohydrate-binding sites present on the monomer.
- Protein Structure Section, Macromolecular Crystallography Laboratory, National Cancer Institute-Frederick, Frederick, MD 21702-1201, USA.
Organizational Affiliation: