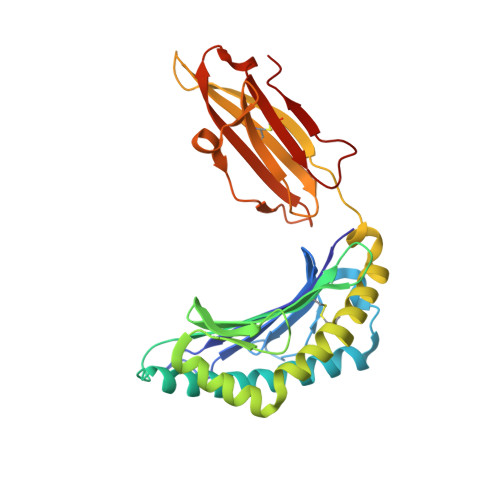
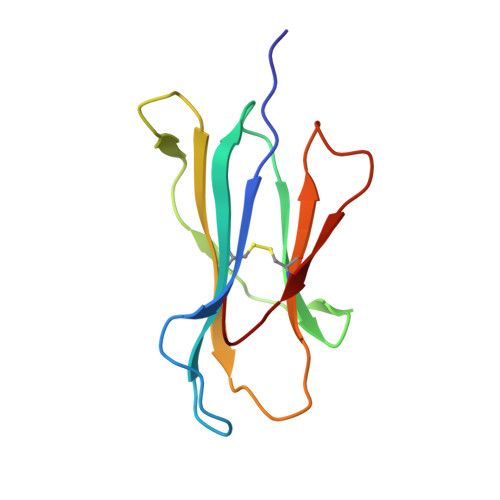
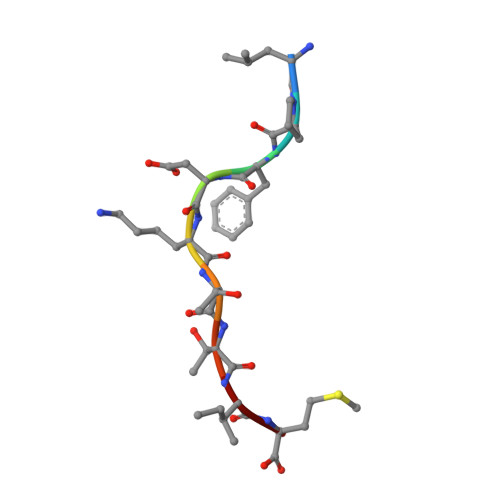
Cross-reactive CD8+ T-cell immunity between the pandemic H1N1-2009 and H1N1-1918 influenza A viruses.
Gras, S., Kedzierski, L., Valkenburg, S.A., Laurie, K., Liu, Y.C., Denholm, J.T., Richards, M.J., Rimmelzwaan, G.F., Kelso, A., Doherty, P.C., Turner, S.J., Rossjohn, J., Kedzierska, K.(2010) Proc Natl Acad Sci U S A 107: 12599-12604
- PubMed: 20616031
- DOI: https://doi.org/10.1073/pnas.1007270107
- Primary Citation of Related Structures:
3LKN, 3LKO, 3LKP, 3LKQ, 3LKR, 3LKS - PubMed Abstract:
Preexisting T-cell immunity directed at conserved viral regions promotes enhanced recovery from influenza virus infections, with there being some evidence of cross-protection directed at variable peptides. Strikingly, many of the immunogenic peptides derived from the current pandemic A(H1N1)-2009 influenza virus are representative of the catastrophic 1918 "Spanish flu" rather than more recent "seasonal" strains. We present immunological and structural analyses of cross-reactive CD8(+) T-cell-mediated immunity directed at a variable (although highly cross-reactive) immunodominant NP(418-426) peptide that binds to a large B7 family (HLA-B*3501/03/0702) found throughout human populations. Memory CD8(+) T-cell specificity was probed for 12 different NP(418) mutants that emerged over the 9 decades between the 1918 and 2009 pandemics. Although there is evidence of substantial cross-reactivity among seasonal NP(418) mutants, current memory T-cell profiles show no preexisting immunity to the 2009-NP(418) variant or the 1918-NP(418) variant. Natural infection with the A(H1N1)-2009 virus, however, elicits CD8(+) T cells specific for the 2009-NP(418) and 1918-NP(418) epitopes. This analysis points to the potential importance of cross-reactive T-cell populations that cover the possible spectrum of T-cell variants and suggests that the identification of key residues/motifs that elicit cross-reactive T-cell sets could facilitate the evolution of immunization protocols that provide a measure of protection against unpredicted pandemic influenza viruses. Thus, it is worth exploring the potential of vaccines that incorporate peptide variants with a proven potential for broader immunogenicity, especially to those that are not recognized by the current memory T-cell pool generated by exposure to influenza variants that cause successive seasonal epidemics.
- Protein Crystallography Unit, Department of Biochemistry and Molecular Biology, Monash University, Clayton, Victoria 3800, Australia.
Organizational Affiliation: