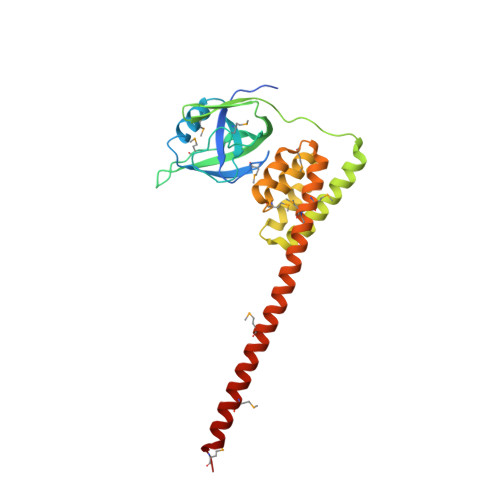
Structure of the N-terminal fragment of Escherichia coli Lon protease
Li, M., Gustchina, A., Rasulova, F.S., Melnikov, E.E., Maurizi, M.R., Rotanova, T.V., Dauter, Z., Wlodawer, A.(2010) Acta Crystallogr D Biol Crystallogr 66: 865-873
- PubMed: 20693685
- DOI: https://doi.org/10.1107/S0907444910019554
- Primary Citation of Related Structures:
3LJC - PubMed Abstract:
The structure of a recombinant construct consisting of residues 1-245 of Escherichia coli Lon protease, the prototypical member of the A-type Lon family, is reported. This construct encompasses all or most of the N-terminal domain of the enzyme. The structure was solved by SeMet SAD to 2.6 A resolution utilizing trigonal crystals that contained one molecule in the asymmetric unit. The molecule consists of two compact subdomains and a very long C-terminal alpha-helix. The structure of the first subdomain (residues 1-117), which consists mostly of beta-strands, is similar to that of the shorter fragment previously expressed and crystallized, whereas the second subdomain is almost entirely helical. The fold and spatial relationship of the two subdomains, with the exception of the C-terminal helix, closely resemble the structure of BPP1347, a 203-amino-acid protein of unknown function from Bordetella parapertussis, and more distantly several other proteins. It was not possible to refine the structure to satisfactory convergence; however, since almost all of the Se atoms could be located on the basis of their anomalous scattering the correctness of the overall structure is not in question. The structure reported here was also compared with the structures of the putative substrate-binding domains of several proteins, showing topological similarities that should help in defining the binding sites used by Lon substrates.
- Protein Structure Section, Macromolecular Crystallography Laboratory, National Cancer Institute at Frederick, Frederick, MD 21702-1201, USA.
Organizational Affiliation: