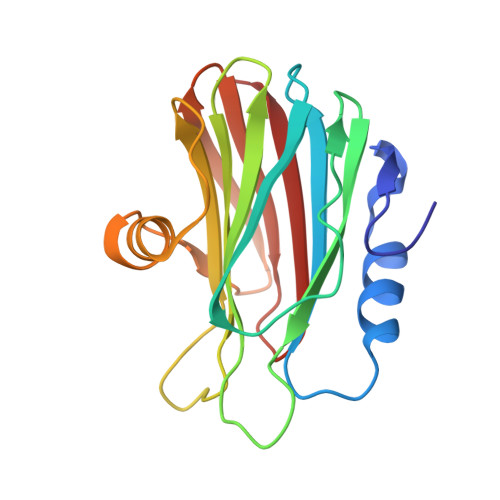
Structural insights into the oligomerization and architecture of eukaryotic membrane pore-forming toxins.
Mechaly, A.E., Bellomio, A., Gil-Carton, D., Morante, K., Valle, M., Gonzalez-Manas, J.M., Guerin, D.M.(2011) Structure 19: 181-191
- PubMed: 21300287
- DOI: https://doi.org/10.1016/j.str.2010.11.013
- Primary Citation of Related Structures:
3LIM - PubMed Abstract:
Pore-forming toxins (PFTs) are proteins that are secreted as soluble molecules and are inserted into membranes to form oligomeric transmembrane pores. In this paper, we report the crystal structure of Fragaceatoxin C (FraC), a PFT isolated from the sea anemone Actinia fragacea, at 1.8 Å resolution. It consists of a crown-shaped nonamer with an external diameter of about 11.0 nm and an internal diameter of approximately 5.0 nm. Cryoelectron microscopy studies of FraC in lipid bilayers reveal the pore structure that traverses the membrane. The shape and dimensions of the crystallographic oligomer are fully consistent with the membrane pore. The FraC structure provides insight into the interactions governing the assembly process and suggests the structural changes that allow for membrane insertion. We propose a nonameric pore model that spans the membrane by forming a lipid-free α-helical bundle pore.
- Unidad de Biofísica (CSIC-UPV/EHU), Universidad del País Vasco, Barrio Sarriena S/N, 48940, Leioa, Vizcaya, Spain.
Organizational Affiliation: