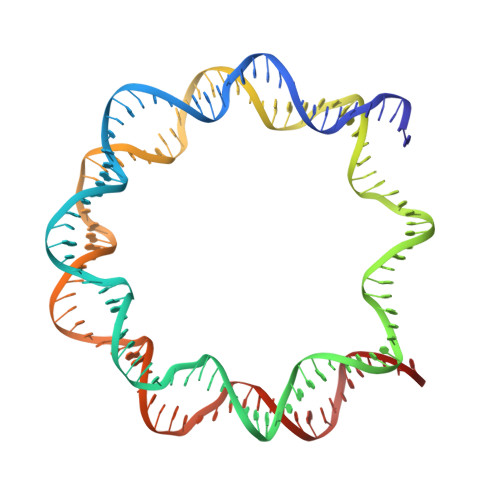
Structural insight into the sequence dependence of nucleosome positioning
Wu, B., Mohideen, K., Vasudevan, D., Davey, C.A.(2010) Structure 18: 528-536
- PubMed: 20399189
- DOI: https://doi.org/10.1016/j.str.2010.01.015
- Primary Citation of Related Structures:
3LEL - PubMed Abstract:
Nucleosome positioning displays sequence dependency and contributes to genomic regulation in a site-specific manner. We solved the structures of nucleosome core particle composed of strong positioning TTTAA elements flanking the nucleosome center. The positioning strength of the super flexible TA dinucleotide is consistent with its observed central location within minor groove inward regions, where it can contribute maximally to energetically challenging minor groove bending, kinking and compression. The marked preference for TTTAA and positioning power of the site 1.5 double helix turns from the nucleosome center relates to a unique histone protein motif at this location, which enforces a sustained, extremely narrow minor groove via a hydrophobic "sugar clamp." Our analysis sheds light on the basis of nucleosome positioning and indicates that the histone octamer has evolved not to fully minimize sequence discrimination in DNA binding.
- Division of Structural and Computational Biology, School of Biological Sciences, Nanyang Technological University, 60 Nanyang Drive, Singapore 637551.
Organizational Affiliation: