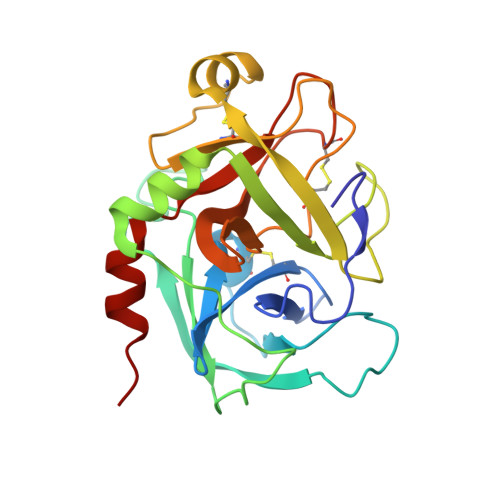
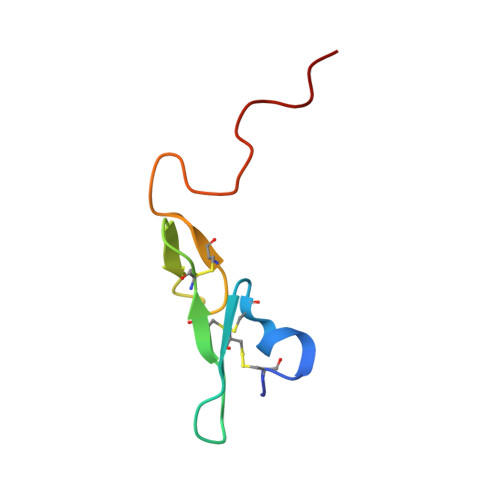
Studies of Benzothiophene Template as Potent Factor IXa (FIXa) Inhibitors in Thrombosis.
Wang, S., Beck, R., Blench, T., Burd, A., Buxton, S., Malic, M., Ayele, T., Shaikh, S., Chahwala, S., Chander, C., Holland, R., Merette, S., Zhao, L., Blackney, M., Watts, A.(2010) J Med Chem 53: 1465-1472
- PubMed: 20121198
- DOI: https://doi.org/10.1021/jm901475e
- Primary Citation of Related Structures:
3LC3 - PubMed Abstract:
FIXa is a serine protease enzyme involved in the intrinsic pathway of the coagulation cascade. The upstream intervention of the coagulation cascade in selectively inhibiting FIXa would leave hemostasis intact via the extrinsic pathway, leading to an optimum combination of efficacy and safety with low incidence of bleeding. We have identified 2-amindinobenzothiophene template as a lead scaffold for FIXa inhibiton based on its homology with urokinase plasminogen activator (uPA). Subsequent SAR work on the template revealed a number of highly potent FIXa inhibitors, though with moderate selectivity against FXa. X-ray study with one of the analogues demonstrated active site binding interaction with the induced opening of the S1 beta pocket and a secondary binding at the S2-S4 sites, which is in direct contrast with the previous finding.
- Department of Medicinal Chemistry, Trigen Ltd., Emmanuel Kaye Building,1B Manresa Road, London SW3 6LR, UK. shouming_wang@yahoo.co.uk
Organizational Affiliation: