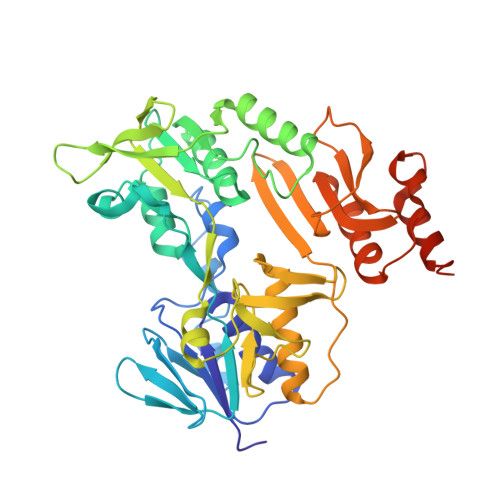
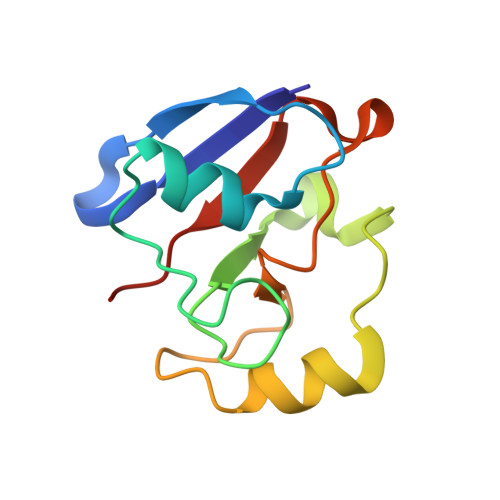
Crystal structure of the putidaredoxin reductase x putidaredoxin electron transfer complex.
Sevrioukova, I.F., Poulos, T.L., Churbanova, I.Y.(2010) J Biological Chem 285: 13616-13620
- PubMed: 20179327
- DOI: https://doi.org/10.1074/jbc.M110.104968
- Primary Citation of Related Structures:
3LB8 - PubMed Abstract:
In the camphor monooxygenase system from Pseudomonas putida, the [2Fe-2S]-containing putidaredoxin (Pdx) shuttles electrons between the NADH-dependent putidaredoxin reductase (Pdr) and cytochrome P450(cam). The mechanism of the Pdr.Pdx redox couple has been investigated by a variety of techniques. One of the exceptions is x-ray crystallography as the native partners associate weakly and resist co-crystallization. Here, we present the 2.6-A x-ray structure of a catalytically active complex between Pdr and Pdx C73S/C85S chemically cross-linked via the Lys(409Pdr)-Glu(72Pdx) pair. The 365 A(2) Pdr-Pdx interface is predominantly hydrophobic with one central Arg(310Pdr)-Asp(38Pdx) salt bridge, likely assisting docking and orienting the partners optimally for electron transfer, and a few peripheral hydrogen bonds. A predicted 12-A-long electron transfer route between FAD and [2Fe-2S] includes flavin flanking Trp(330Pdr) and the iron ligand Cys(39Pdx). The x-ray model agrees well with the experimental and theoretical results and suggests that the linked Pdx must undergo complex movements during turnover to accommodate P450(cam), which could limit the Pdx-to-P450(cam) electron transfer reaction.
- Department of Molecular Biology and Biochemistry, University of California, Irvine, California 92697-3900, USA. sevrioui@uci.edu
Organizational Affiliation: