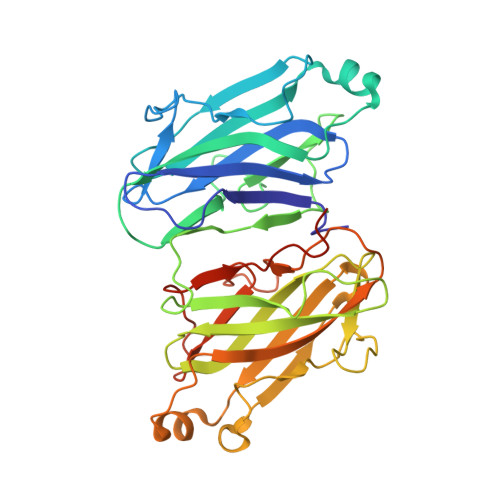
Modular Arrangement of a Cellulosomal Scaffoldin Subunit Revealed from the Crystal Structure of a Cohesin Dyad
Noach, I., Levy-Assaraf, M., Lamed, R., Shimon, L.J.W., Frolow, F., Bayer, E.A.(2010) J Mol Biology 399: 294-305
- PubMed: 20394754
- DOI: https://doi.org/10.1016/j.jmb.2010.04.013
- Primary Citation of Related Structures:
3L8Q - PubMed Abstract:
The cellulosome complex is composed of a conglomerate of subunits, each of which comprises a set of interacting functional modules. Scaffoldin (Sca), a major cellulosomal subunit, is responsible for organizing the cellulolytic subunits into the complex. This is accomplished by the interaction of two complementary classes of modules-a cohesin (Coh) module on the Sca subunit and a dockerin module on each of the enzymatic subunits. Although individual Coh modules from different cellulosomal scaffoldins have been subjected to intensive structural investigation, the Sca subunit in its entirety has not, and there remains a paucity of information on the arrangement and interactions of Cohs within the Sca subunit. In the present work, we describe the crystal structure of a type II Coh dyad from the ScaB "adaptor" Sca of Acetivibrio cellulolyticus. The ScaB Cohs are oriented in an "antiparallel" manner relative to one another, with their dockerin-interacting surfaces (beta-strands 8-3-6-5) facing the same direction-aligned on the same plane. A set of extensive hydrophobic and hydrogen-bond contacts between the Cohs and the short interconnecting linker segment between them stabilizes the modular orientation. This Coh dyad structure provides novel information about Coh-Coh association and arrangement in the Sca and further insight into intermodular linker interactions. Putative structural arrangements of a hexamodular complex, composed of the Coh dyad bound to two X-dockerin modules, were suggested.
- Department of Biological Chemistry, The Weizmann Institute of Science, Rehovot 76100, Israel.
Organizational Affiliation: