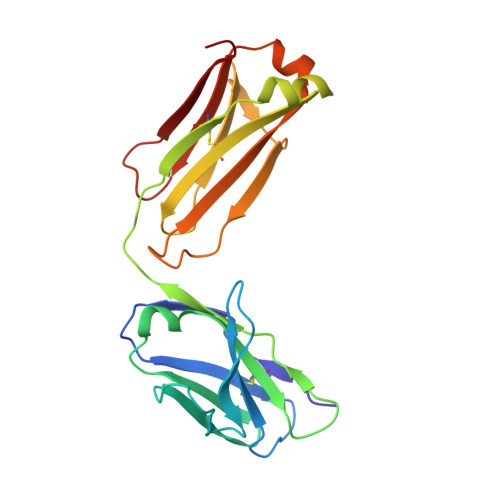
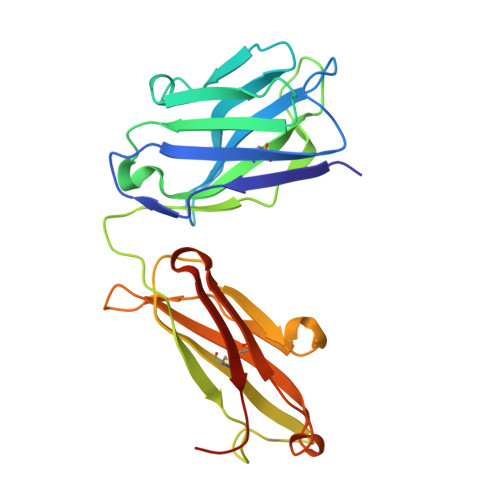
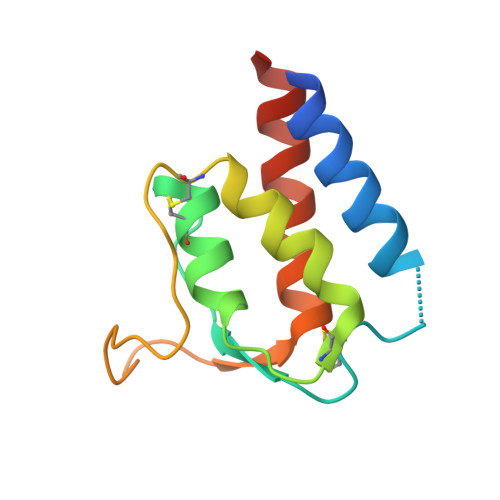
Human framework adaptation of a mouse anti-human IL-13 antibody.
Fransson, J., Teplyakov, A., Raghunathan, G., Chi, E., Cordier, W., Dinh, T., Feng, Y., Giles-Komar, J., Gilliland, G., Lollo, B., Malia, T.J., Nishioka, W., Obmolova, G., Zhao, S., Zhao, Y., Swanson, R.V., Almagro, J.C.(2010) J Mol Biology 398: 214-231
- PubMed: 20226193
- DOI: https://doi.org/10.1016/j.jmb.2010.03.004
- Primary Citation of Related Structures:
3L5W, 3L5X, 3L7F, 4PS4 - PubMed Abstract:
Humanization of a potent neutralizing mouse anti-human IL-13 antibody (m836) using a method called human framework adaptation (HFA) is reported. HFA consists of two steps: human framework selection (HFS) and specificity-determining residue optimization (SDRO). The HFS step involved generation of a library of m836 antigen binding sites combined with diverse human germline framework regions (FRs), which were selected based on structural and sequence similarities between mouse variable domains and a repertoire of human antibody germline genes. SDRO consisted of diversifying specificity-determining residues and selecting variants with improved affinity using phage display. HFS of m836 resulted in a 5-fold loss of affinity, whereas SDRO increased the affinity up to 100-fold compared to the HFS antibody. Crystal structures of Fabs in complex with IL-13 were obtained for m836, the HFS variant chosen for SDRO, and one of the highest-affinity SDRO variants. Analysis of the structures revealed that major conformational changes in FR-H1 and FR-H3 occurred after FR replacement, but none of them had an evident direct impact on residues in contact with IL-13. Instead, subtle changes affected the V(L)/V(H) (variable-light domain/variable-heavy domain) interface and were likely responsible for the 5-fold decreased affinity. After SDRO, increased affinity resulted mainly from rearrangements in hydrogen-bonding pattern at the antibody/antigen interface. Comparison with m836 putative germline genes suggested interesting analogies between natural affinity maturation and the engineering process that led to the potent HFA anti-human IL-13 antibody.
- Centocor R&D, Inc., 3210 Merryfield Row, San Diego, CA 92121, USA.
Organizational Affiliation: