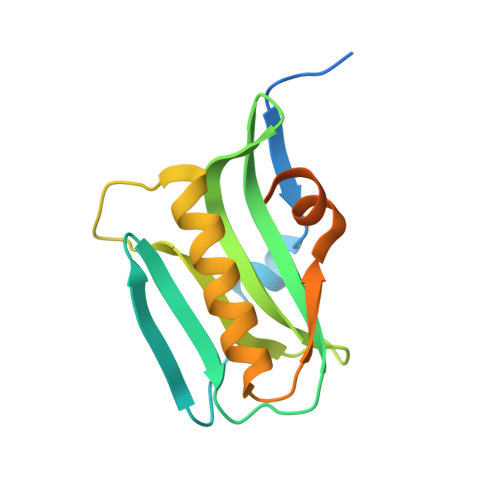
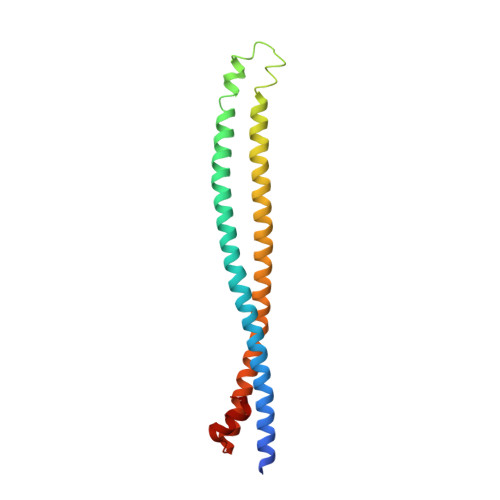
Structural insights into phosphoinositide 3-kinase activation by the influenza A virus NS1 protein
Hale, B.G., Kerry, P.S., Jackson, D., Precious, B.L., Gray, A., Killip, M.J., Randall, R.E., Russell, R.J.(2010) Proc Natl Acad Sci U S A 107: 1954-1959
- PubMed: 20133840
- DOI: https://doi.org/10.1073/pnas.0910715107
- Primary Citation of Related Structures:
3L4Q - PubMed Abstract:
Seasonal epidemics and periodic worldwide pandemics caused by influenza A viruses are of continuous concern. The viral nonstructural (NS1) protein is a multifunctional virulence factor that antagonizes several host innate immune defenses during infection. NS1 also directly stimulates class IA phosphoinositide 3-kinase (PI3K) signaling, an essential cell survival pathway commonly mutated in human cancers. Here, we present a 2.3-A resolution crystal structure of the NS1 effector domain in complex with the inter-SH2 (coiled-coil) domain of p85beta, a regulatory subunit of PI3K. Our data emphasize the remarkable isoform specificity of this interaction, and provide insights into the mechanism by which NS1 activates the PI3K (p85beta:p110) holoenzyme. A model of the NS1:PI3K heterotrimeric complex reveals that NS1 uses the coiled-coil as a structural tether to sterically prevent normal inhibitory contacts between the N-terminal SH2 domain of p85beta and the p110 catalytic subunit. Furthermore, in this model, NS1 makes extensive contacts with the C2/kinase domains of p110, and a small acidic alpha-helix of NS1 sits adjacent to the highly basic activation loop of the enzyme. During infection, a recombinant influenza A virus expressing NS1 with charge-disruption mutations in this acidic alpha-helix is unable to stimulate the production of phosphatidylinositol 3,4,5-trisphosphate or the phosphorylation of Akt. Despite this, the charge-disruption mutations in NS1 do not affect its ability to interact with the p85beta inter-SH2 domain in vitro. Overall, these data suggest that both direct binding of NS1 to p85beta (resulting in repositioning of the N-terminal SH2 domain) and possible NS1:p110 contacts contribute to PI3K activation.
- Biomedical Science Research Complex, University of St Andrews, North Haugh, St Andrews, Fife, KY16 9ST, United Kingdom.
Organizational Affiliation: