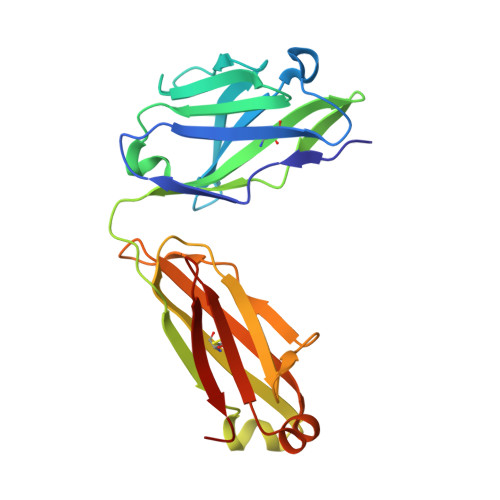
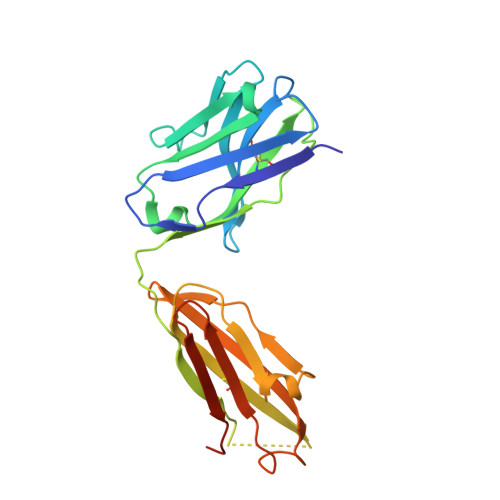
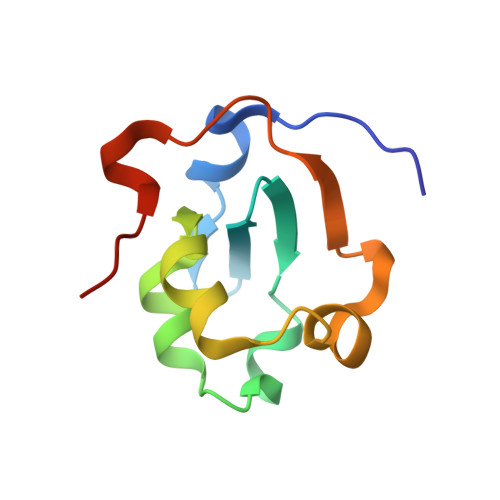
Structural evidence for the functional importance of the heme domain mobility in flavocytochrome b2.
Diep Le, K.H., Lederer, F., Golinelli-Pimpaneau, B.(2010) J Mol Biology 400: 518-530
- PubMed: 20546754
- DOI: https://doi.org/10.1016/j.jmb.2010.05.035
- Primary Citation of Related Structures:
3KS0 - PubMed Abstract:
Yeast flavocytochrome b(2) (Fcb2) is an L-lactate:cytochrome c oxidoreductase in the mitochondrial intermembrane space participating in cellular respiration. Each enzyme subunit consists of a cytochrome b(5)-like heme domain and a flavodehydrogenase (FDH) domain. In the Fcb2 crystal structure, the heme domain is mobile relative to the tetrameric FDH core in one out of two subunits. The monoclonal antibody B2B4, elicited against the holoenzyme, recognizes only the native heme domain in the holoenzyme. When bound, it suppresses the intramolecular electron transfer from flavin to heme b(2), hence cytochrome c reduction. We report here the crystal structure of the heme domain in complex with the Fab at 2.7 A resolution. The Fab epitope on the heme domain includes the two exposed propionate groups of the heme, which are hidden in the interface between the domains in the complete subunit. The structure discloses an unexpected plasticity of Fcb2 in the neighborhood of the heme cavity, in which the heme has rotated. The epitope overlaps with the docking area of the FDH domain onto the heme domain, indicating that the antibody displaces the heme domain in a movement of large amplitude. We suggest that the binding sites on the heme domain of cytochrome c and of the FDH domain also overlap and therefore that cytochrome c binding also requires the heme domain to move away from the FDH domain, so as to allow electron transfer between the two hemes. Based on this hypothesis, we propose a possible model of the Fcb2.cytochrome c complex. Interestingly, this model shares similarity with that of the cytochrome b(5) x cytochrome c complex, in which cytochrome c binds to the surface around the exposed heme edge of cytochrome b(5). The present results therefore support the idea that the heme domain mobility is an inherent component of the Fcb2 functioning.
- Laboratoire d'Enzymologie et Biochimie Structurales, CNRS, 1 avenue de la Terrasse, 91198 Gif-sur-Yvette, France.
Organizational Affiliation: