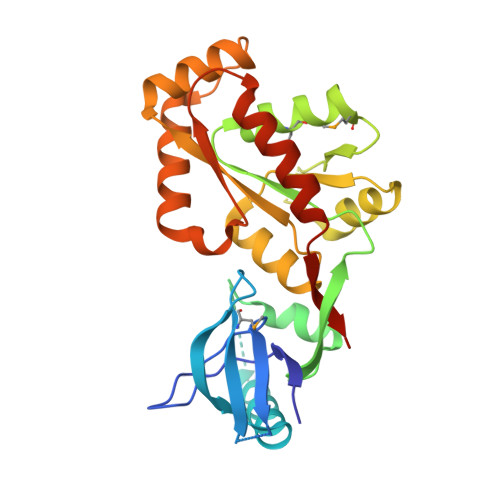
Crystal structure of the SH3-kinase fragment of tight junction protein 3 (TJP3) in apo-form
Tong, Y., Nedyalkova, L., Tempel, W., Zhong, N., Guan, X., Arrowsmith, C.H., Edwards, A.M., Bountra, C., Weigelt, J., Bochkarev, A., Park, H.To be published.