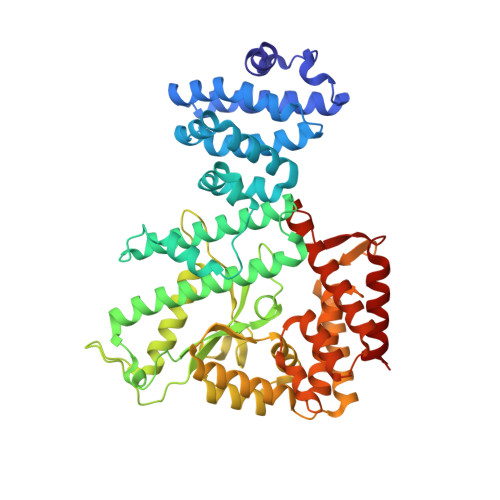
Structure of the Adenylylation Domain of E. coli Glutamine Synthetase Adenylyl Transferase: Evidence for Gene Duplication and Evolution of a New Active Site.
Xu, Y., Carr, P.D., Vasudevan, S.G., Ollis, D.L.(2010) J Mol Biology 396: 773-784
- PubMed: 20026075
- DOI: https://doi.org/10.1016/j.jmb.2009.12.011
- Primary Citation of Related Structures:
3K7D - PubMed Abstract:
The X-ray structure of the C-terminal fragment, containing residues 449-946, of Escherichia coli glutamine synthetase adenylyl transferase (ATase) has been determined. ATase is part of the cascade that regulates the enzymatic activity of E. coli glutamine synthetase, a key component of the cell's machinery for the uptake of ammonia. It has two enzymatic activities, adenylyl removase (AR) and adenylyl transferase (AT), which are located in distinct catalytic domains that are separated by a regulatory (R) domain. We previously reported the three-dimensional structure of the AR domain (residues 1-440). The present structure contains both the R and AT domains. AR and AT share 24% sequence identity and also contain the beta-polymerase motif that is characteristic of many nucleotidylyl transferase enzymes. The structures overlap with an rmsd of 2.4 A when the superhelical R domain is omitted. A model for the complete ATase molecule is proposed, along with some refinements of domain boundaries. A rather more speculative model for the complex of ATase with glutamine synthetase and the nitrogen signal transduction protein PII is also presented.
- Structural Biology Division, The Walter and Eliza Hall Institute of Medical Research, Parkville, Australia.
Organizational Affiliation: