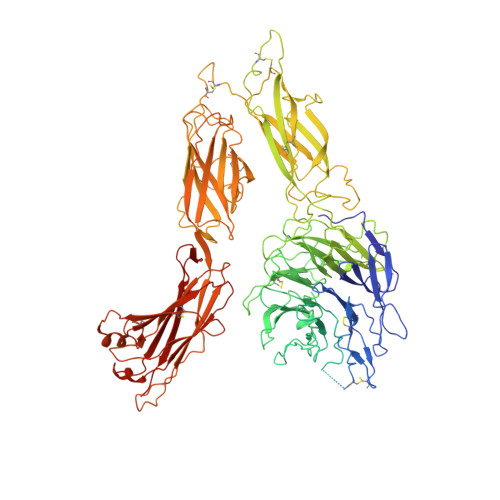
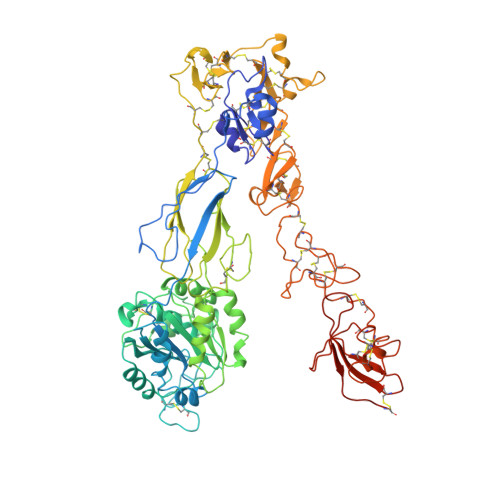
Structure of an integrin with an alphaI domain, complement receptor type 4.
Xie, C., Zhu, J., Chen, X., Mi, L., Nishida, N., Springer, T.A.(2010) EMBO J 29: 666-679
- PubMed: 20033057
- DOI: https://doi.org/10.1038/emboj.2009.367
- Primary Citation of Related Structures:
3K6S, 3K71, 3K72 - PubMed Abstract:
We report the structure of an integrin with an alphaI domain, alpha(X)beta(2), the complement receptor type 4. It was earlier expected that a fixed orientation between the alphaI domain and the beta-propeller domain in which it is inserted would be required for allosteric signal transmission. However, the alphaI domain is highly flexible, enabling two betaI domain conformational states to couple to three alphaI domain states, and greater accessibility for ligand recognition. Although alpha(X)beta(2) is bent similarly to integrins that lack alphaI domains, the terminal domains of the alpha- and beta-legs, calf-2 and beta-tail, are oriented differently than in alphaI-less integrins. Linkers extending to the transmembrane domains are unstructured. Previous mutations in the beta(2)-tail domain support the importance of extension, rather than a deadbolt, in integrin activation. The locations of further activating mutations and antibody epitopes show the critical role of extension, and conversion from the closed to the open headpiece conformation, in integrin activation. Differences among 10 molecules in crystal lattices provide unprecedented information on interdomain flexibility important for modelling integrin extension and activation.
- Department of Pathology, Harvard Medical School, Immune Disease Institute and Children's Hospital, Boston, MA 02115, USA.
Organizational Affiliation: