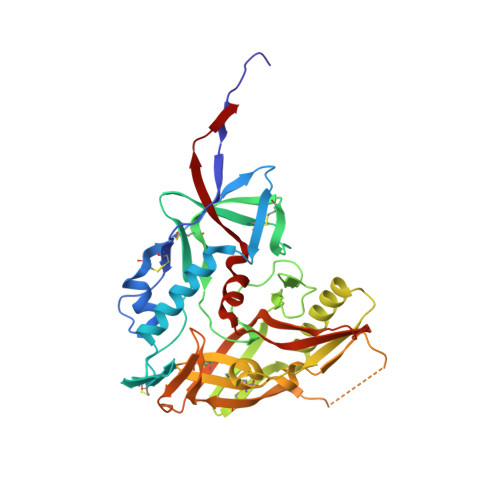
Structure of HIV-1 gp120 with gp41-interactive region reveals layered envelope architecture and basis of conformational mobility.
Pancera, M., Majeed, S., Ban, Y.E., Chen, L., Huang, C.C., Kong, L., Kwon, Y.D., Stuckey, J., Zhou, T., Robinson, J.E., Schief, W.R., Sodroski, J., Wyatt, R., Kwong, P.D.(2010) Proc Natl Acad Sci U S A 107: 1166-1171
- PubMed: 20080564
- DOI: https://doi.org/10.1073/pnas.0911004107
- Primary Citation of Related Structures:
3JWD, 3JWO - PubMed Abstract:
The viral spike of HIV-1 is composed of three gp120 envelope glycoproteins attached noncovalently to three gp41 transmembrane molecules. Viral entry is initiated by binding to the CD4 receptor on the cell surface, which induces large conformational changes in gp120. These changes not only provide a model for receptor-triggered entry, but affect spike sensitivity to drug- and antibody-mediated neutralization. Although some of the details of the CD4-induced conformational change have been visualized by crystal structures and cryoelectron tomograms, the critical gp41-interactive region of gp120 was missing from previous atomic-level characterizations. Here we determine the crystal structure of an HIV-1 gp120 core with intact gp41-interactive region in its CD4-bound state, compare this structure to unliganded and antibody-bound forms to identify structurally invariant and plastic components, and use ligand-oriented cryoelectron tomograms to define component mobility in the viral spike context. Newly defined gp120 elements proximal to the gp41 interface complete a 7-stranded beta-sandwich, which appeared invariant in conformation. Loop excursions emanating from the sandwich form three topologically separate--and structurally plastic--layers, topped off by the highly glycosylated gp120 outer domain. Crystal structures, cryoelectron tomograms, and interlayer chemistry were consistent with a mechanism in which the layers act as a shape-changing spacer, facilitating movement between outer domain and gp41-associated beta-sandwich and providing for conformational diversity used in immune evasion. A "layered" gp120 architecture thus allows movement among alternative glycoprotein conformations required for virus entry and immune evasion, whereas a beta-sandwich clamp maintains gp120-gp41 interaction and regulates gp41 transitions.
- Vaccine Research Center, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD 20892, USA.
Organizational Affiliation: