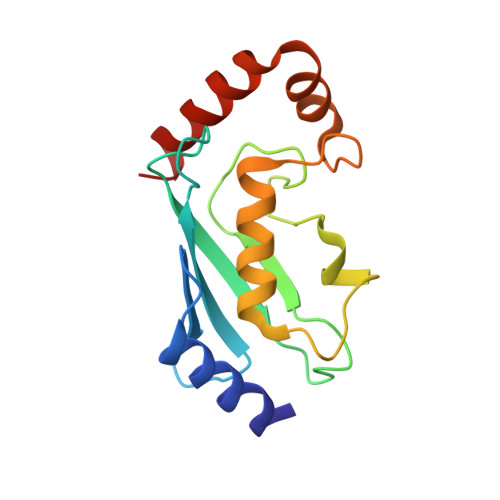
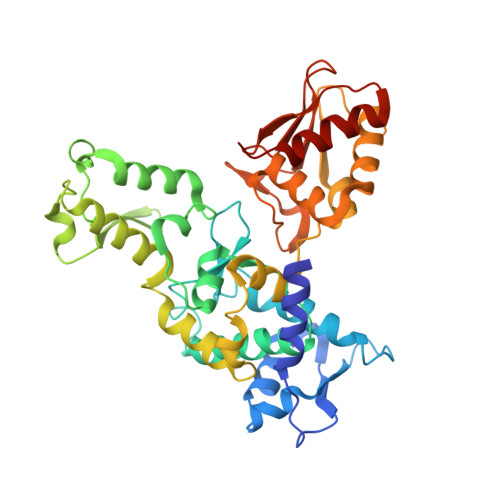
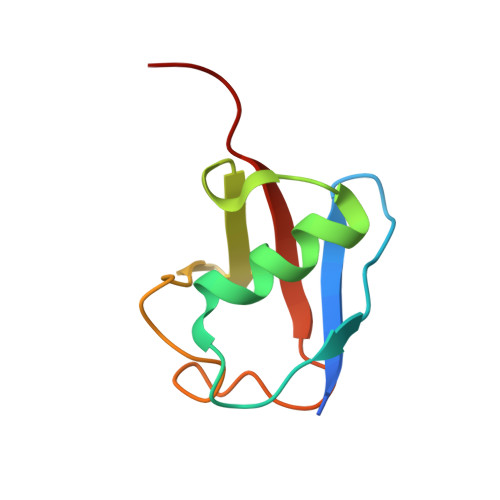
Insights into ubiquitin transfer cascades from a structure of a UbcH5B approximately ubiquitin-HECT(NEDD4L) complex.
Kamadurai, H.B., Souphron, J., Scott, D.C., Duda, D.M., Miller, D.J., Stringer, D., Piper, R.C., Schulman, B.A.(2009) Mol Cell 36: 1095-1102
- PubMed: 20064473
- DOI: https://doi.org/10.1016/j.molcel.2009.11.010
- Primary Citation of Related Structures:
3JVZ, 3JW0 - PubMed Abstract:
In E1-E2-E3 ubiquitin (Ub) conjugation cascades, the E2 first forms a transient E2 approximately Ub covalent complex and then interacts with an E3 for Ub transfer. For cascades involving E3s in the HECT class, Ub is transferred from an associated E2 to the acceptor cysteine in the HECT domain C lobe. To gain insights into this process, we determined the crystal structure of a complex between the HECT domain of NEDD4L and the E2 UbcH5B bearing a covalently linked Ub at its active site (UbcH5B approximately Ub). Noncovalent interactions between UbcH5B and the HECT N lobe and between Ub and the HECT domain C lobe lead to an overall compact structure, with the Ub C terminus sandwiched between UbcH5B and HECT domain active sites. The structure suggests a model for E2-to-HECT Ub transfer, in which interactions between a donor Ub and an acceptor domain constrain upstream and downstream enzymes for conjugation.
- Department of Structural Biology, St. Jude Children's Research Hospital, Memphis, TN 38105, USA.
Organizational Affiliation: