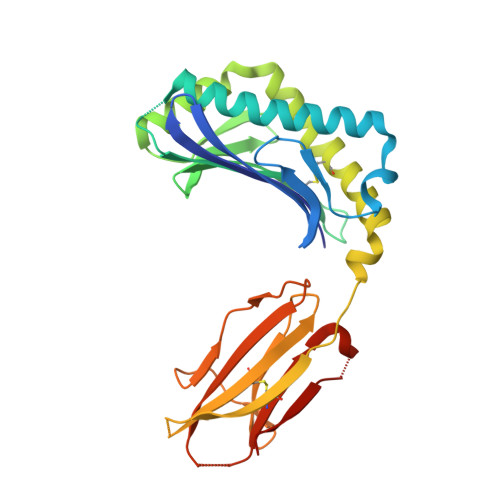
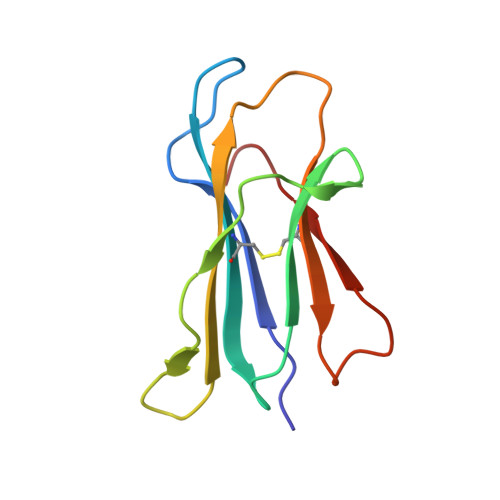
Structural basis for lipid-antigen recognition in avian immunity.
Dvir, H., Wang, J., Ly, N., Dascher, C.C., Zajonc, D.M.(2010) J Immunol 184: 2504-2511
- PubMed: 20100930
- DOI: https://doi.org/10.4049/jimmunol.0903509
- Primary Citation of Related Structures:
3JVG - PubMed Abstract:
CD1 proteins present self- and foreign lipid Ags to activate specific T cells in the mammalian immune system. These T cells play an important role in controlling autoimmune diseases, suppression of tumor growth, and host defense against invading pathogens. Humans use five CD1 isoforms, whereas only two exist in birds. Unlike mammals' CD1, the structure of chicken CD1-2 showed a primitive lipid-binding groove, suggesting that chicken may only recognize single-chain lipids. In contrast, the crystal structure of the second chicken CD1 isoform, chCD1-1, reported in this study at 2.2 A resolution, reveals an elaborated binding groove with a dual-pocket, dual-cleft architecture. The A' and F' deep pockets are separated from each other, but each is connected to a hydrophobic surface cleft, which may participate in lipid binding. The long endogenous ligand found inside the binding groove of chCD1-1, together with binding data on various glycolipids and mycolic acid, strongly suggest that the unique avian CD1 family could bind long dual- and possibly triacyl-chain lipids.
- Department of Cell Biology, La Jolla Institute for Allergy and Immunology, La Jolla CA 92037, USA.
Organizational Affiliation: