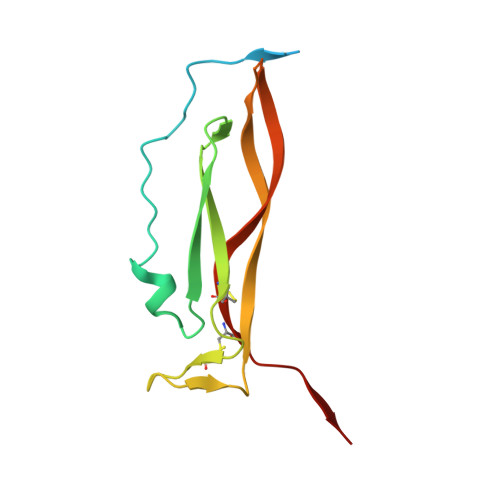
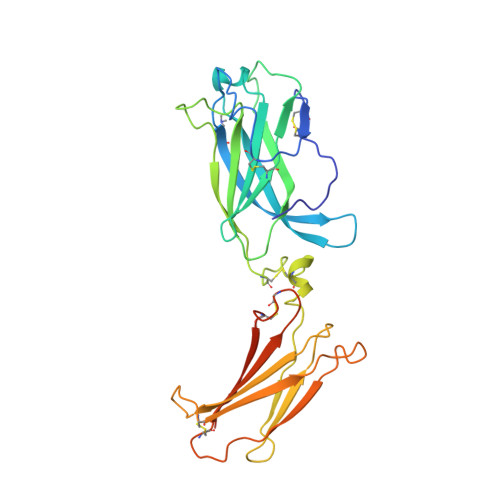
Structural basis of receptor sharing by interleukin 17 cytokines.
Ely, L.K., Fischer, S., Garcia, K.C.(2009) Nat Immunol 10: 1245-1251
- PubMed: 19838198
- DOI: https://doi.org/10.1038/ni.1813
- Primary Citation of Related Structures:
3JVF - PubMed Abstract:
Interleukin 17 (IL-17)-producing helper T cells (T(H)-17 cells), together with their effector cytokines, including members of the IL-17 family, are emerging as key mediators of chronic inflammatory and autoimmune disorders. Here we present the crystal structure of a complex of IL-17 receptor A (IL-17RA) bound to IL-17F in a 1:2 stoichiometry. The mechanism of complex formation was unique for cytokines and involved the engagement of IL-17 by two fibronectin-type domains of IL-17RA in a groove between the IL-17 homodimer interface. Binding of the first receptor to the IL-17 cytokines modulated the affinity and specificity of the second receptor-binding event, thereby promoting heterodimeric versus homodimeric complex formation. IL-17RA used a common recognition strategy to bind to several members of the IL-17 family, which allows it to potentially act as a shared receptor in multiple different signaling complexes.
- Howard Hughes Medical Institute, Department of Molecular and Cellular Physiology, Stanford University School of Medicine, Stanford, California, USA.
Organizational Affiliation: