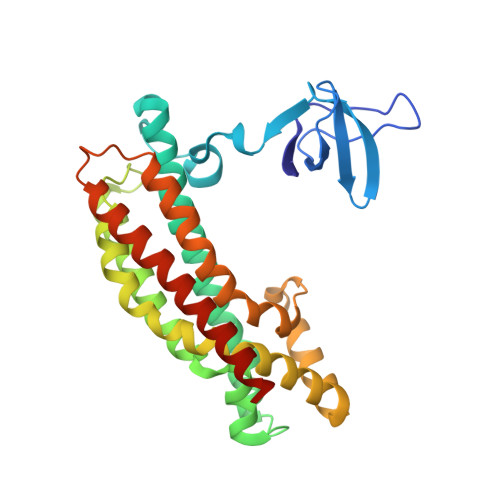
The minimal autoinhibited unit of the guanine nucleotide exchange factor intersectin.
Ahmad, K.F., Lim, W.A.(2010) PLoS One 5: e11291-e11291
- PubMed: 20585582
- DOI: https://doi.org/10.1371/journal.pone.0011291
- Primary Citation of Related Structures:
3JV3 - PubMed Abstract:
Intersectin-1L is a member of the Dbl homology (DH) domain guanine nucleotide exchange factors (GEF) which control Rho-family GTPase signaling. Intersectin-1L is a GEF that is specific for Cdc42. It plays an important role in endocytosis, and is regulated by several partners including the actin regulator N-WASP. Intact intersectin-1L shows low Cdc42 exchange activity, although the isolated catalytic DH domain shows high activity. This finding suggests that the molecule is autoinhibited. To investigate the mechanism of autoinhibition we have constructed a series of domain deletions. We find that the five SH3 domains of intersectin are important for autoinhibition, with the fifth domain (SH3(E)) being sufficient for the bulk of the autoinhibitory effect. This SH3 domain appears to primarily interact with the DH domain. We have determined the crystal structure of the SH3(E)-DH domain construct, which shows a domain swapped arrangement in which the SH3 from one monomer interacts with the DH domain of the other monomer. Analytical ultracentrifugation and gel filtration, however, show that under biochemical concentrations, the construct is fully monomeric. Thus we propose that the actual autoinhibited structure contains the related intramolecular SH3(E)-DH interaction. We propose a model in which this intramolecular interaction may block or distort the GTPase binding region of the DH domain.
- Department of Cellular and Molecular Pharmacology, University of California San Francisco, San Francisco, California, United States of America.
Organizational Affiliation: