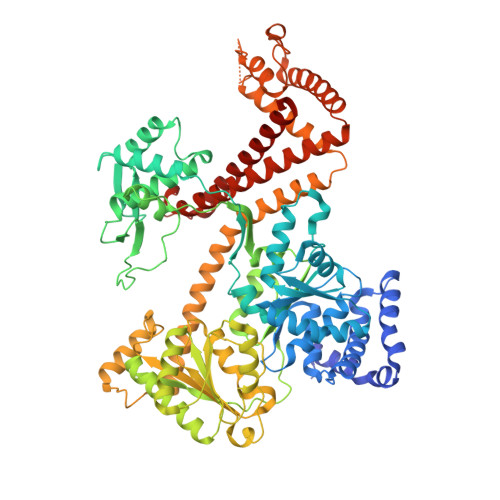
Conformational flexibility and peptide interaction of the translocation ATPase SecA.
Zimmer, J., Rapoport, T.A.(2009) J Mol Biology 394: 606-612
- PubMed: 19850053
- DOI: https://doi.org/10.1016/j.jmb.2009.10.024
- Primary Citation of Related Structures:
3JUX, 3JV2 - PubMed Abstract:
The SecA ATPase forms a functional complex with the protein-conducting SecY channel to translocate polypeptides across the bacterial cell membrane. SecA recognizes the translocation substrate and catalyzes its unidirectional movement through the SecY channel. The recent crystal structure of the Thermotoga maritima SecA-SecYEG complex shows the ATPase in a conformation where the nucleotide-binding domains (NBDs) have closed around a bound ADP-BeFx complex and SecA's polypeptide-binding clamp is shut. Here, we present the crystal structure of T. maritima SecA in isolation, determined in its ADP-bound form at 3.1 A resolution. SecA alone has a drastically different conformation in which the nucleotide-binding pocket between NBD1 and NBD2 is open and the preprotein cross-linking domain has rotated away from both NBDs, thereby opening the polypeptide-binding clamp. To investigate how this clamp binds polypeptide substrates, we also determined a structure of Bacillus subtilis SecA in complex with a peptide at 2.5 A resolution. This structure shows that the peptide augments the highly conserved beta-sheet at the back of the clamp. Taken together, these structures suggest a mechanism by which ATP hydrolysis can lead to polypeptide translocation.
- Howard Hughes Medical Institute and Department of Cell Biology, Harvard Medical School, 240 Longwood Avenue, Boston, MA 02115, USA.
Organizational Affiliation: