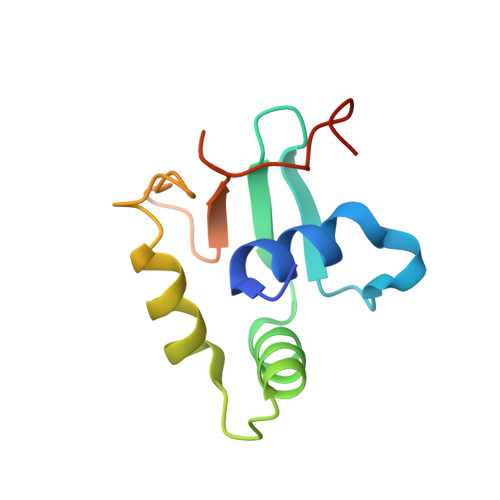
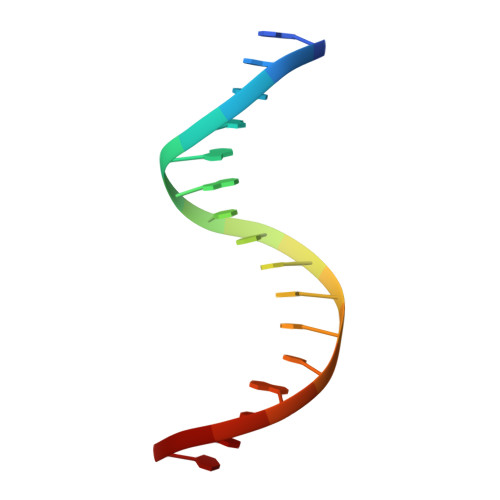
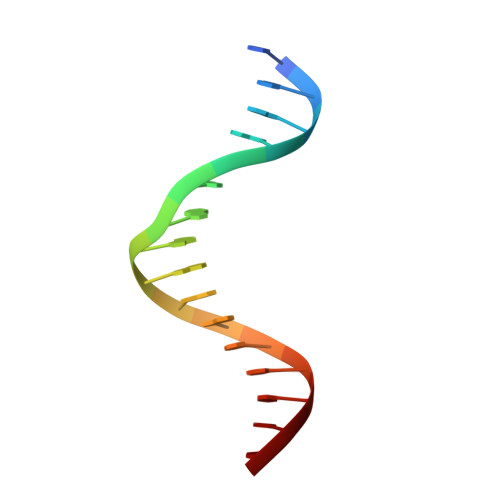
Crystal structure of mouse Elf3 C-terminal DNA-binding domain in complex with type II TGF-beta receptor promoter DNA.
Agarkar, V.B., Babayeva, N.D., Wilder, P.J., Rizzino, A., Tahirov, T.H.(2010) J Mol Biology 397: 278-289
- PubMed: 20079749
- DOI: https://doi.org/10.1016/j.jmb.2010.01.017
- Primary Citation of Related Structures:
3JTG - PubMed Abstract:
The Ets family of transcription factors is composed of more than 30 members. One of its members, Elf3, is expressed in virtually all epithelial cells as well as in many tumors, including breast tumors. Several studies observed that the promoter of the type II TGF-beta receptor gene (TbetaR-II) is strongly stimulated by Elf3 via two adjacent Elf3 binding sites, the A-site and the B-site. Here, we report the 2.2 A resolution crystal structure of a mouse Elf3 C-terminal fragment, containing the DNA-binding Ets domain, in complex with the B-site of mouse type II TGF-beta receptor promoter DNA (mTbetaR-II(DNA)). Elf3 contacts the core GGAA motif of the B-site from a major groove similar to that of known Ets proteins. However, unlike other Ets proteins, Elf3 also contacts sequences of the A-site from the minor groove of the DNA. DNA binding experiments and cell-based transcription studies indicate that minor groove interaction by Arg349 located in the Ets domain is important for Elf3 function. Equally interesting, previous studies have shown that the C-terminal region of Elf3, which flanks the Ets domain, is required for Elf3 binding to DNA. In this study, we determined that Elf3 amino acid residues within this flanking region, including Trp361, are important for the structural integrity of the protein as well as for the Efl3 DNA binding and transactivation activity.
- Eppley Institute for Research in Cancer and Allied Diseases, University of Nebraska Medical Center, 987696 Nebraska Medical Center, Omaha, NE 68198-7696, USA.
Organizational Affiliation: