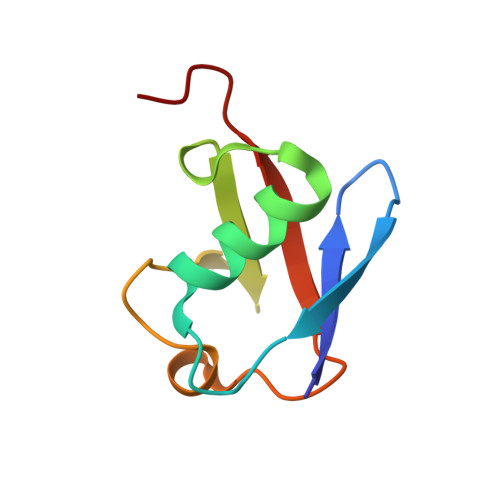
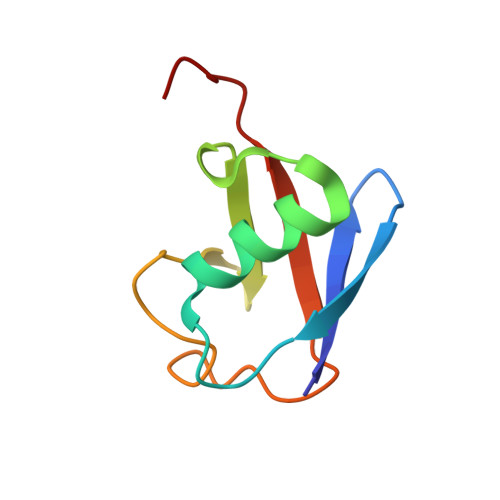
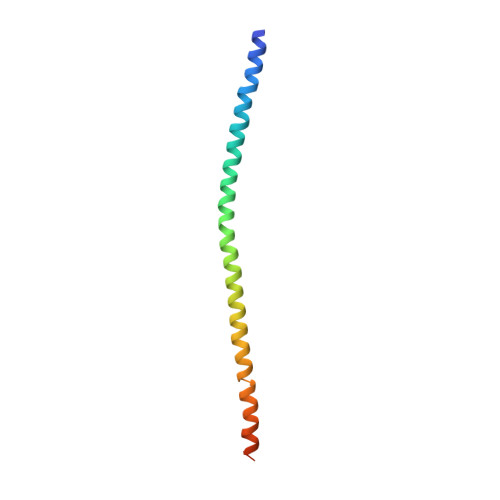Crystal structure of the NEMO ubiquitin-binding domain in complex with Lys 63-linked di-ubiquitin
Yoshikawa, A., Sato, Y., Yamashita, M., Mimura, H., Yamagata, A., Fukai, S.(2009) FEBS Lett 583: 3317-3322
- PubMed: 19766637
- DOI: https://doi.org/10.1016/j.febslet.2009.09.028
- Primary Citation of Related Structures:
3JSV - PubMed Abstract:
NEMO is essential for activation of the NF-kappaB signaling pathway, which is regulated by ubiquitination of proteins. The C-terminal leucine zipper of NEMO and its adjacent coiled-coil region (CC2-LZ) reportedly bind to linear ubiquitin chains with 1 microM affinity and to Lys 63-linked chains with 100 microM affinity. Here we report the crystal structure of the CC2-LZ region of mouse NEMO in complex with Lys 63-linked di-ubiquitin (K63-Ub(2)) at 2.7A resolution. The ubiquitin-binding region consists of a 130A-long helix and forms a parallel coiled-coil dimer. The Ile 44-centered hydrophobic patch of ubiquitin is recognized in the middle of the NEMO ubiquitin-binding region. NEMO interacts with each K63-Ub(2)via a single ubiquitin-binding site, consistent with low affinity binding with K63-Ub(2).
- Structural Biology Laboratory, Life Science Division, Synchrotron Radiation Research Organization and Institute of Molecular and Cellular Biosciences, The University of Tokyo, Tokyo 113-0032, Japan.
Organizational Affiliation: