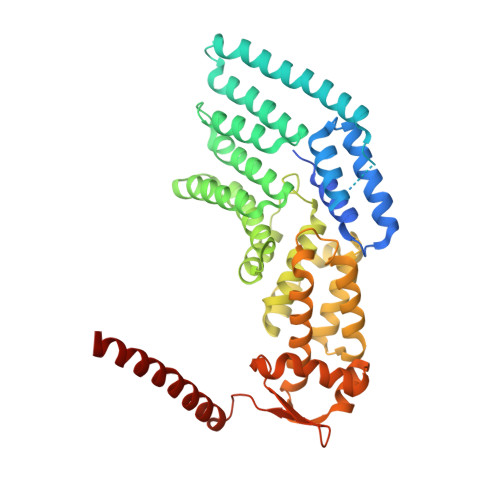
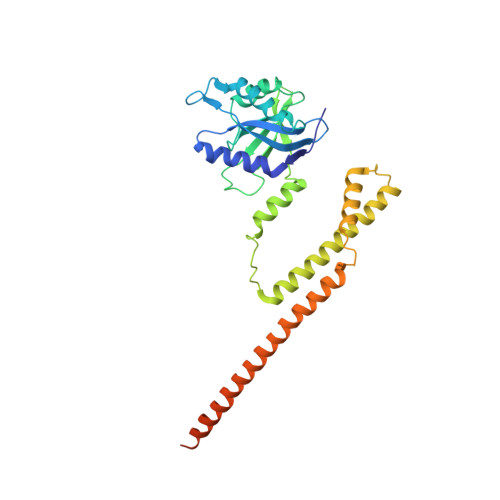
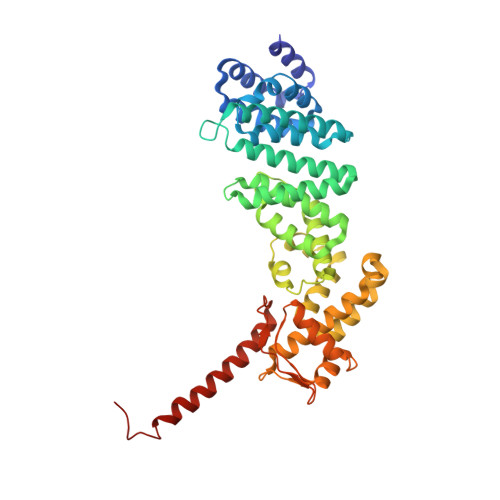
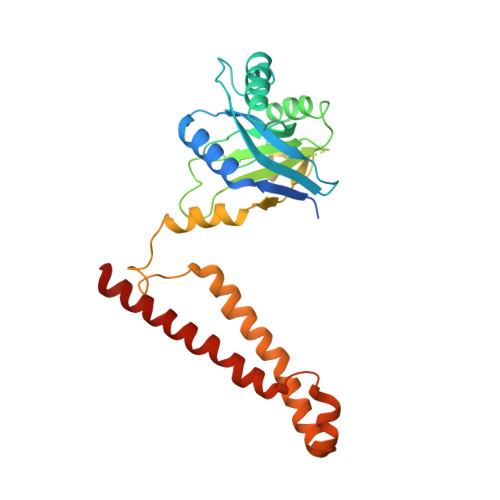
Atomic structure of the 26S proteasome lid reveals the mechanism of deubiquitinase inhibition.
Dambacher, C.M., Worden, E.J., Herzik, M.A., Martin, A., Lander, G.C.(2016) Elife 5: e13027-e13027
- PubMed: 26744777
- DOI: https://doi.org/10.7554/eLife.13027
- Primary Citation of Related Structures:
3JCK - PubMed Abstract:
The 26S proteasome is responsible for the selective, ATP-dependent degradation of polyubiquitinated cellular proteins. Removal of ubiquitin chains from targeted substrates at the proteasome is a prerequisite for substrate processing and is accomplished by Rpn11, a deubiquitinase within the 'lid' sub-complex. Prior to the lid's incorporation into the proteasome, Rpn11 deubiquitinase activity is inhibited to prevent unwarranted deubiquitination of polyubiquitinated proteins. Here we present the atomic model of the isolated lid sub-complex, as determined by cryo-electron microscopy at 3.5 Å resolution, revealing how Rpn11 is inhibited through its interaction with a neighboring lid subunit, Rpn5. Through mutagenesis of specific residues, we describe the network of interactions that are required to stabilize this inhibited state. These results provide significant insight into the intricate mechanisms of proteasome assembly, outlining the substantial conformational rearrangements that occur during incorporation of the lid into the 26S holoenzyme, which ultimately activates the deubiquitinase for substrate degradation.
- Department of Integrative Structural and Computational Biology, The Scripps Research Institute, La Jolla, United States.
Organizational Affiliation: