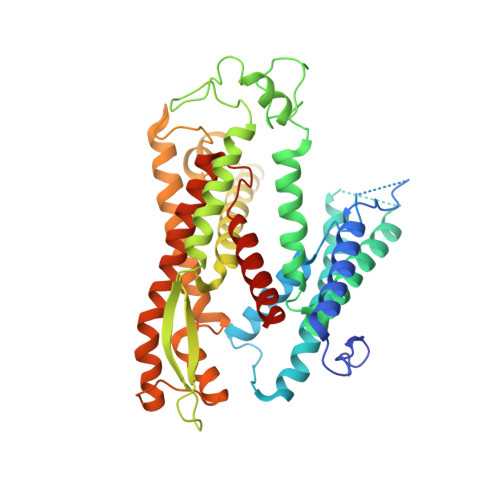
Structure of the Sec61 channel opened by a signal sequence.
Voorhees, R.M., Hegde, R.S.(2016) Science 351: 88-91
- PubMed: 26721998
- DOI: https://doi.org/10.1126/science.aad4992
- Primary Citation of Related Structures:
3JC2 - PubMed Abstract:
Secreted and integral membrane proteins compose up to one-third of the biological proteome. These proteins contain hydrophobic signals that direct their translocation across or insertion into the lipid bilayer by the Sec61 protein-conducting channel. The molecular basis of how hydrophobic signals within a nascent polypeptide trigger channel opening is not understood. Here, we used cryo-electron microscopy to determine the structure of an active Sec61 channel that has been opened by a signal sequence. The signal supplants helix 2 of Sec61α, which triggers a rotation that opens the central pore both axially across the membrane and laterally toward the lipid bilayer. Comparisons with structures of Sec61 in other states suggest a pathway for how hydrophobic signals engage the channel to gain access to the lipid bilayer.
- MRC Laboratory of Molecular Biology, Medical Research Council, Francis Crick Avenue, Cambridge CB2 0QH, UK.
Organizational Affiliation: