Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM.
Li, X., Mooney, P., Zheng, S., Booth, C.R., Braunfeld, M.B., Gubbens, S., Agard, D.A., Cheng, Y.(2013) Nat Methods 10: 584-590
- PubMed: 23644547
- DOI: https://doi.org/10.1038/nmeth.2472
- Primary Citation of Related Structures:
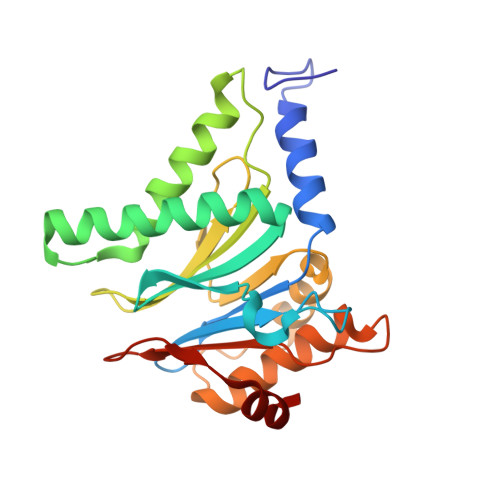
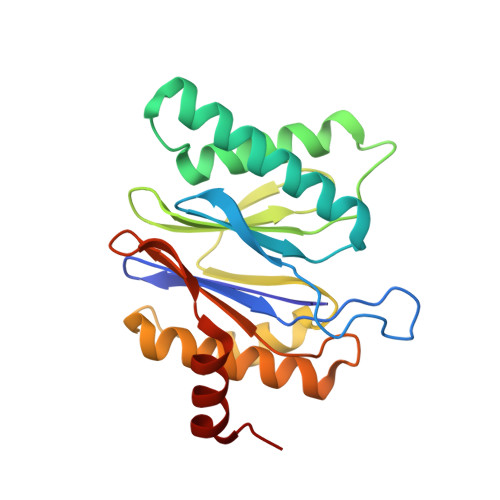
3J9I - PubMed Abstract:
In recent work with large high-symmetry viruses, single-particle electron cryomicroscopy (cryo-EM) has achieved the determination of near-atomic-resolution structures by allowing direct fitting of atomic models into experimental density maps. However, achieving this goal with smaller particles of lower symmetry remains challenging. Using a newly developed single electron-counting detector, we confirmed that electron beam-induced motion substantially degrades resolution, and we showed that the combination of rapid readout and nearly noiseless electron counting allow image blurring to be corrected to subpixel accuracy, restoring intrinsic image information to high resolution (Thon rings visible to ∼3 Å). Using this approach, we determined a 3.3-Å-resolution structure of an ∼700-kDa protein with D7 symmetry, the Thermoplasma acidophilum 20S proteasome, showing clear side-chain density. Our method greatly enhances image quality and data acquisition efficiency-key bottlenecks in applying near-atomic-resolution cryo-EM to a broad range of protein samples.
- The Keck Advanced Microscopy Laboratory, Department of Biochemistry and Biophysics, University of California, San Francisco (UCSF), San Francisco, California, USA.
Organizational Affiliation: