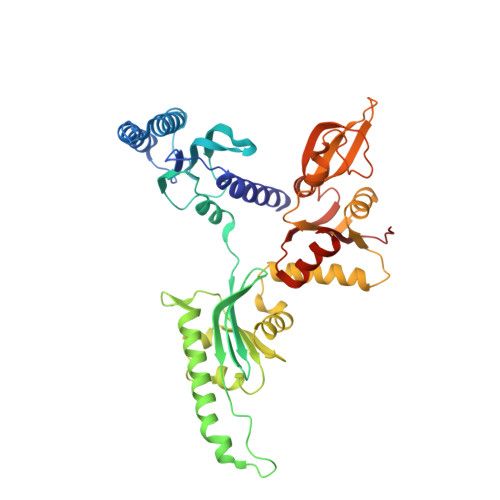
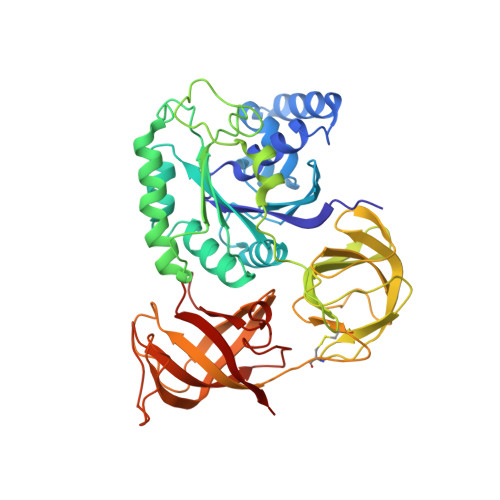
Structure of the mammalian ribosomal pre-termination complex associated with eRF1*eRF3*GDPNP.
des Georges, A., Hashem, Y., Unbehaun, A., Grassucci, R.A., Taylor, D., Hellen, C.U., Pestova, T.V., Frank, J.(2014) Nucleic Acids Res 42: 3409-3418
- PubMed: 24335085
- DOI: https://doi.org/10.1093/nar/gkt1279
- Primary Citation of Related Structures:
3J5Y - PubMed Abstract:
Eukaryotic translation termination results from the complex functional interplay between two release factors, eRF1 and eRF3, in which GTP hydrolysis by eRF3 couples codon recognition with peptidyl-tRNA hydrolysis by eRF1. Here, we present a cryo-electron microscopy structure of pre-termination complexes associated with eRF1•eRF3•GDPNP at 9.7 -Å resolution, which corresponds to the initial pre-GTP hydrolysis stage of factor attachment and stop codon recognition. It reveals the ribosomal positions of eRFs and provides insights into the mechanisms of stop codon recognition and triggering of eRF3's GTPase activity.
- Howard Hughes Medical Institute, Chevy Chase, MD, USA, Department of Biochemistry and Molecular Biophysics, Columbia University, New York, NY, USA, Department of Cell Biology, SUNY Downstate Medical Center, Brooklyn, NY, USA, Department of Pharmacology, Case Western Reserve University, Cleveland, OH, USA and Department of Biological Sciences, Columbia University, New York, NY, USA.
Organizational Affiliation: