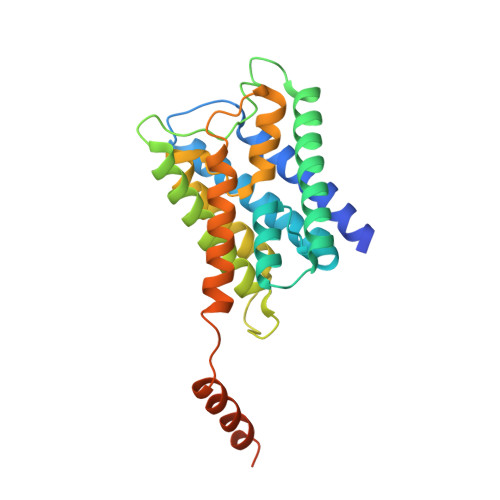
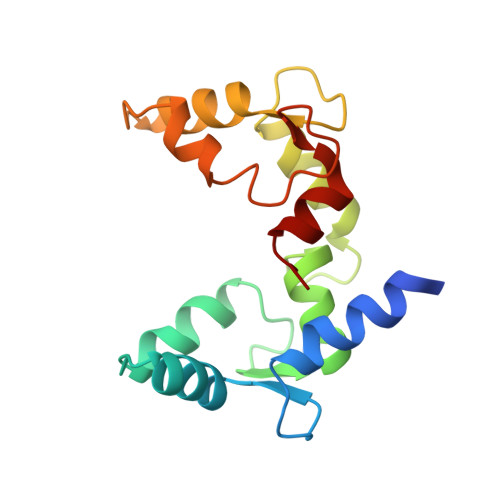
Allosteric mechanism of water-channel gating by Ca(2+)-calmodulin.
Reichow, S.L., Clemens, D.M., Freites, J.A., Nemeth-Cahalan, K.L., Heyden, M., Tobias, D.J., Hall, J.E., Gonen, T.(2013) Nat Struct Mol Biol 20: 1085-1092
- PubMed: 23893133
- DOI: https://doi.org/10.1038/nsmb.2630
- Primary Citation of Related Structures:
3J41 - PubMed Abstract:
Calmodulin (CaM) is a universal regulatory protein that communicates the presence of calcium to its molecular targets and correspondingly modulates their function. This key signaling protein is important for controlling the activity of hundreds of membrane channels and transporters. However, understanding of the structural mechanisms driving CaM regulation of full-length membrane proteins has remained elusive. In this study, we determined the pseudoatomic structure of full-length mammalian aquaporin-0 (AQP0, Bos taurus) in complex with CaM, using EM to elucidate how this signaling protein modulates water-channel function. Molecular dynamics and functional mutation studies reveal how CaM binding inhibits AQP0 water permeability by allosterically closing the cytoplasmic gate of AQP0. Our mechanistic model provides new insight, only possible in the context of the fully assembled channel, into how CaM regulates multimeric channels by facilitating cooperativity between adjacent subunits.
- 1] Howard Hughes Medical Institute, Janelia Farm Research Campus, Ashburn, Virginia, USA. [2].
Organizational Affiliation: