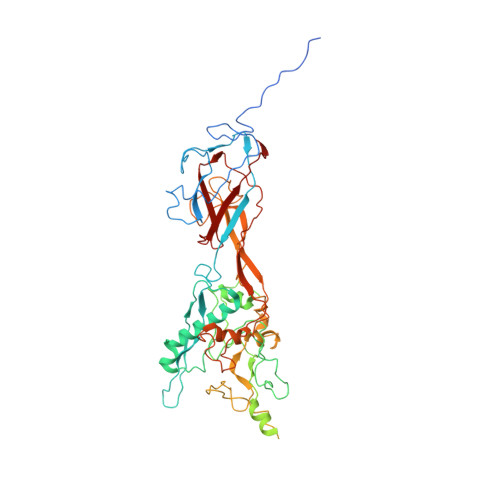
Model of the Trimeric Fiber and Its Interactions with the Pentameric Penton Base of Human Adenovirus by Cryo-electron Microscopy.
Liu, H., Wu, L., Zhou, Z.H.(2011) J Mol Biology 406: 764-774
- PubMed: 21146538
- DOI: https://doi.org/10.1016/j.jmb.2010.11.043
- Primary Citation of Related Structures:
3IZO - PubMed Abstract:
Adenovirus invades host cells by first binding to host receptors through a trimeric fiber, which contains three domains: a receptor-binding knob domain, a long flexible shaft domain, and a penton base-attachment tail domain. Although the structure of the knob domain associated with a portion of the shaft has been solved by X-ray crystallography, the in situ structure of the fiber in the virion is not known; thus, it remains a mystery how the trimeric fiber attaches to its underlying pentameric penton base. By high-resolution cryo-electron microscopy, we have determined the structure of the human adenovirus type 5 (Ad5) to 3.6-Å resolution and have reported the full atomic models for its capsid proteins, but not for the fiber whose density cannot be directly interpreted due to symmetry mismatch with the penton base. Here, we report the determination of the Ad5 fiber structure and its mode of attachment to the pentameric penton base by using an integrative approach of multi-resolution filtering, homology modeling, computational simulation of mismatched symmetries, and fitting of atomic models into cryo-electron microscopy density maps. Our structure reveals that the interactions between the trimeric fiber and the pentameric penton base are mediated by a hydrophobic ring on the top surface of the penton base and three flexible tails inserted into three of the five available grooves formed by neighboring subunits of penton base. These interaction sites provide the molecular basis for the symmetry mismatch and can be targeted for optimizing adenovirus for gene therapy applications.
- Department of Microbiology, Immunology and Molecular Genetics, University of California, Los Angeles, Los Angeles, CA 90095-7364, USA.
Organizational Affiliation: