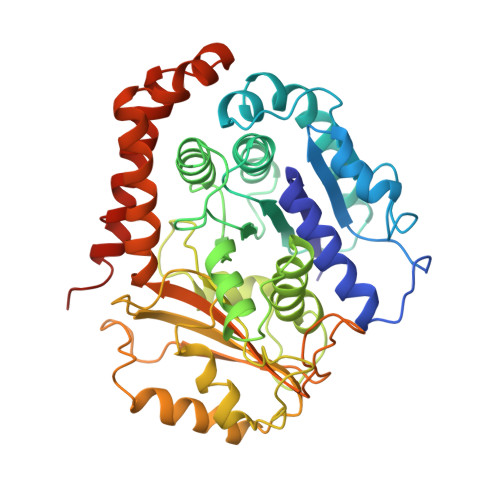
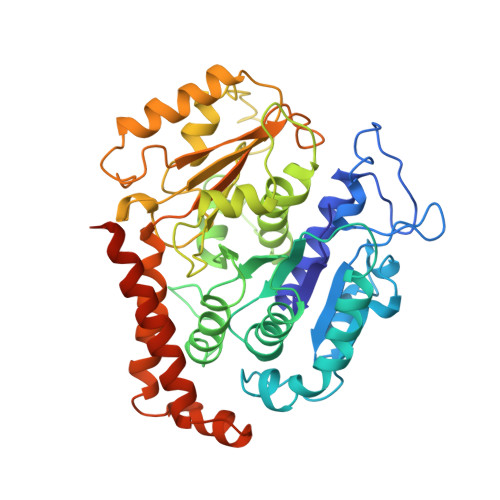
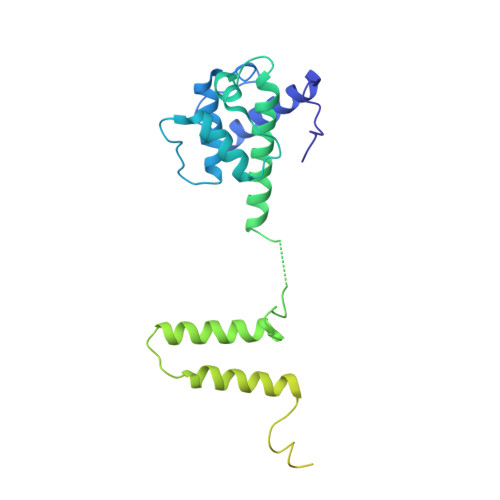
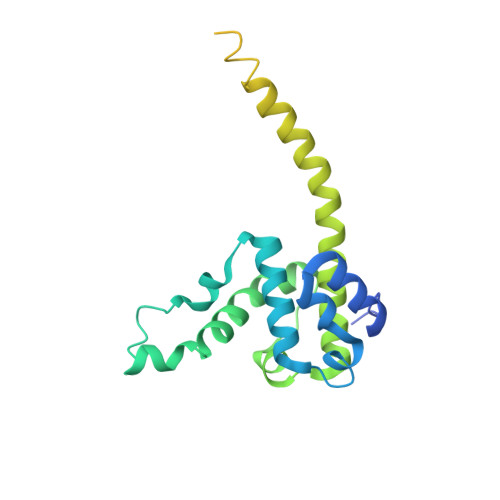
The Ndc80 kinetochore complex forms oligomeric arrays along microtubules.
Alushin, G.M., Ramey, V.H., Pasqualato, S., Ball, D.A., Grigorieff, N., Musacchio, A., Nogales, E.(2010) Nature 467: 805-810
- PubMed: 20944740
- DOI: https://doi.org/10.1038/nature09423
- Primary Citation of Related Structures:
3IZ0 - PubMed Abstract:
The Ndc80 complex is a key site of regulated kinetochore-microtubule attachment (a process required for cell division), but the molecular mechanism underlying its function remains unknown. Here we present a subnanometre-resolution cryo-electron microscopy reconstruction of the human Ndc80 complex bound to microtubules, sufficient for precise docking of crystal structures of the component proteins. We find that the Ndc80 complex binds the microtubule with a tubulin monomer repeat, recognizing α- and β-tubulin at both intra- and inter-tubulin dimer interfaces in a manner that is sensitive to tubulin conformation. Furthermore, Ndc80 complexes self-associate along protofilaments through interactions mediated by the amino-terminal tail of the NDC80 protein, which is the site of phospho-regulation by Aurora B kinase. The complex's mode of interaction with the microtubule and its oligomerization suggest a mechanism by which Aurora B could regulate the stability of load-bearing kinetochore-microtubule attachments.
- Biophysics Graduate Group, University of California, Berkeley, California 94720, USA.
Organizational Affiliation: