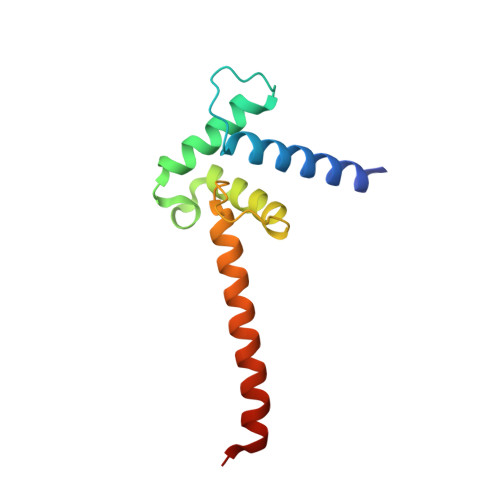
Structure of the Taz2 domain of p300: insights into ligand binding.
Miller, M., Dauter, Z., Cherry, S., Tropea, J.E., Wlodawer, A.(2009) Acta Crystallogr D Biol Crystallogr 65: 1301-1308
- PubMed: 19966416
- DOI: https://doi.org/10.1107/S0907444909040153
- Primary Citation of Related Structures:
3IO2 - PubMed Abstract:
CBP and its paralog p300 are histone acetyl transferases that regulate gene expression by interacting with multiple transcription factors via specialized domains. The structure of a segment of human p300 protein (residues 1723-1836) corresponding to the extended zinc-binding Taz2 domain has been investigated. The crystal structure was solved by the SAD approach utilizing the anomalous diffraction signal of the bound Zn ions. The structure comprises an atypical helical bundle stabilized by three Zn ions and closely resembles the solution structures determined previously for shorter peptides. Residues 1813-1834 from the current construct form a helical extension of the C-terminal helix and make extensive crystal-contact interactions with the peptide-binding site of Taz2, providing additional insights into the mechanism of the recognition of diverse transactivation domains (TADs) by Taz2. On the basis of these results and molecular modeling, a hypothetical model of the binding of phosphorylated p53 TAD1 to Taz2 has been proposed.
- Protein Structure Section, Macromolecular Crystallography Laboratory, NCI-Frederick, Frederick, Maryland 21702-1201, USA. mariami@mail.nih.gov
Organizational Affiliation: