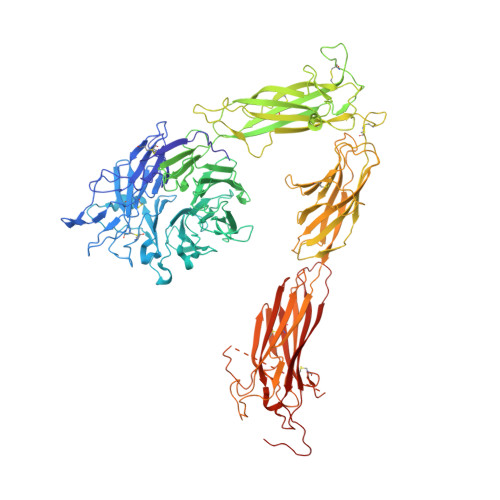
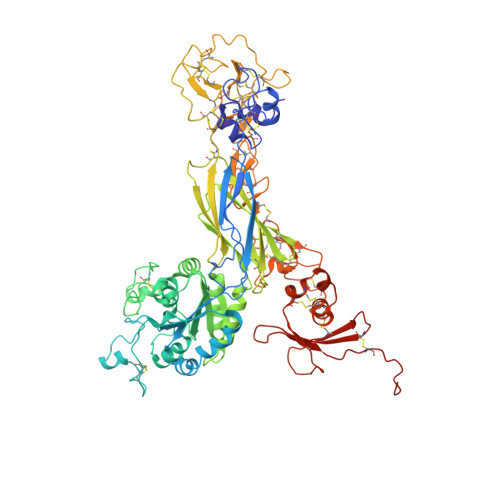
Crystal structure of the complete integrin alphaVbeta3 ectodomain plus an alpha/beta transmembrane fragment.
Xiong, J.P., Mahalingham, B., Alonso, J.L., Borrelli, L.A., Rui, X., Anand, S., Hyman, B.T., Rysiok, T., Muller-Pompalla, D., Goodman, S.L., Arnaout, M.A.(2009) J Cell Biol 186: 589-600
- PubMed: 19704023
- DOI: https://doi.org/10.1083/jcb.200905085
- Primary Citation of Related Structures:
3IJE - PubMed Abstract:
We determined the crystal structure of 1TM-alphaVbeta3, which represents the complete unconstrained ectodomain plus short C-terminal transmembrane stretches of the alphaV and beta3 subunits. 1TM-alphaVbeta3 is more compact and less active in solution when compared with DeltaTM-alphaVbeta3, which lacks the short C-terminal stretches. The structure reveals a bent conformation and defines the alpha-beta interface between IE2 (EGF-like 2) and the thigh domains. Modifying this interface by site-directed mutagenesis leads to robust integrin activation. Fluorescent lifetime imaging microscopy of inactive full-length alphaVbeta3 on live cells yields a donor-membrane acceptor distance, which is consistent with the bent conformation and does not change in the activated integrin. These data are the first direct demonstration of conformational coupling of the integrin leg and head domains, identify the IE2-thigh interface as a critical steric barrier in integrin activation, and suggest that inside-out activation in intact cells may involve conformational changes other than the postulated switch to a genu-linear state.
- Program in Leukocyte Biology and Inflammation, Massachusetts General Hospital, Harvard Medical School, Charlestown, MA 02129, USA.
Organizational Affiliation: