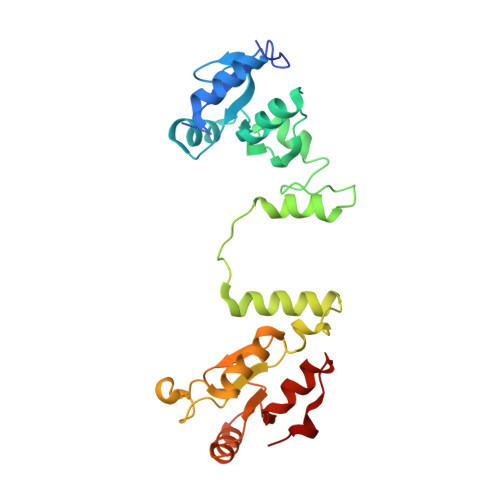
Structural and functional interaction between the human DNA repair proteins DNA ligase IV and XRCC4
Wu, P.Y., Frit, P., Meesala, S., Dauvillier, S., Modesti, M., Andres, S.N., Huang, Y., Sekiguchi, J., Calsou, P., Salles, B., Junop, M.S.(2009) Mol Cell Biol 11: 3163-3172
- PubMed: 19332554
- DOI: https://doi.org/10.1128/MCB.01895-08
- Primary Citation of Related Structures:
3II6 - PubMed Abstract:
Nonhomologous end-joining represents the major pathway used by human cells to repair DNA double-strand breaks. It relies on the XRCC4/DNA ligase IV complex to reseal DNA strands. Here we report the high-resolution crystal structure of human XRCC4 bound to the carboxy-terminal tandem BRCT repeat of DNA ligase IV. The structure differs from the homologous Saccharomyces cerevisiae complex and reveals an extensive DNA ligase IV binding interface formed by a helix-loop-helix structure within the inter-BRCT linker region, as well as significant interactions involving the second BRCT domain, which induces a kink in the tail region of XRCC4. We further demonstrate that interaction with the second BRCT domain of DNA ligase IV is necessary for stable binding to XRCC4 in cells, as well as to achieve efficient dominant-negative effects resulting in radiosensitization after ectopic overexpression of DNA ligase IV fragments in human fibroblasts. Together our findings provide unanticipated insight for understanding the physical and functional architecture of the nonhomologous end-joining ligation complex.
- CNRS, IPBS (Institut de Pharmacologie et de Biologie Structurale), Toulouse, France.
Organizational Affiliation: